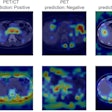
A deep-learning algorithm that can prescreen chest radiographs for an incorrectly positioned peripherally inserted central catheter (PICC) was presented at the recent Society for Imaging Informatics in Medicine (SIIM) annual meeting. The algorithm can alert radiologists to immediately prioritize those cases.
A team from Massachusetts General Hospital (MGH) developed an artificial intelligence (AI) platform based on deep learning that can identify the location of the PICC line tip on chest radiographs. The model could predict the PICC tip line within an average of 3 mm of the actual location, and it returns results in a little over a second, according to presenter Hyunkwang Lee, a doctoral student.
"This system [could] be implemented directly on the imaging device or as part of the PACS to triage the 'correct' and 'incorrect' cases," Lee said. The paper received the SIIM 2017 New Investigator Travel Award.
PICC positioning
A PICC is inserted into a vein in the patient's arm and threaded near the heart to provide intravenous access for administration of medicine, fluid, and chemotherapy drugs. An incorrectly positioned PICC can lead to serious complications, so the final PICC location is confirmed after placement on a chest radiograph. Radiologists have a high accuracy rate for interpreting the location of the PICC on these studies, but delays in interpreting the cases can be significant, Lee said.
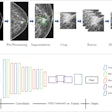
As a result, the MGH team developed a fully automated, deep learning-based algorithm for PICC line detection. The system includes a preprocessing module for image quality normalization, a PICC detection AI algorithm for segmentation of the region of interest, and a postprocessing module to filter out false-positive results and provide accurate localization of the PICC tip, Lee said.
To test the algorithm, the researchers retrospectively collected a dataset of 800 DICOM radiographs: 600 were used for training and validation of the deep-learning model and 200 were reserved for testing. The chest x-ray images were characterized by low image quality, as well as wide variation in pixel contrast, the locations of internal and external artifacts, and patient positioning, Lee said. These images also had high resolution, with an average size of 2,801 x 3,195 pixels.
"So we definitely need the preprocessing module to normalize this image quality," he said.
The preprocessing module applies a bilateral filter for denoising and edge enhancement. Adaptive histogram equalization is then performed to normalize image contrast, he said.
AI detection
Once preprocessing is complete, the image is then ready for the second step -- AI detection of the PICC. The group initially used a "patch"-based approach with a deep convolutional neural network (AlexNet, pretrained on the ImageNet image database) to perform this task. Images were spliced into smaller image patches, which were manually annotated as containing one of 10 classes: background, vertebral body, electrocardiogram (ECG), shoulder, lung, other lines, PICC, rib, tissue, and other object. Sample patches of these 10 classes were used to train the system.
Once the network finishes classifying the patches and completely segments the entire image, it generates a PICC image "mask" to identify the significant pixels in the image patches that had been classified as a PICC. A postprocessing module is then applied in the final step in the process. False-positive PICC results are pruned with a Hough line transform algorithm, Lee said.
"A smoothly carved PICC line trajectory can [then] be generated by merging the significant nearby contours and to compute the PICC line tip location," he said.
The final result is an image that has been annotated with the trajectory of the PICC and the tip location, according to the group.
Performance
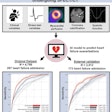
The researchers then calculated the performance of the model by measuring the distance from the predicted PICC line tip location to the ground truth. They noticed, however, that the patch-based approach had a critical drawback that would hinder its user in a clinical setting.
"A patch-based approach requires redundant pixel-wide computations, especially for the larger-sized images," Lee said. With an average of over 58,000 patches per image, that took almost a minute and a half to perform.
To improve computational efficiency, the researchers elected to replace the patch-based approach with a fully convolutional neural network, which "can be trained end to end, pixels to pixels, with input images and their corresponding pixel-wide title labels," Lee said.
As a result, the fully convolutional neural network can classify the entire image at once, rather than having to first process individual image patches, according to the group. This change yielded significant performance improvements.
| Comparison of AI approaches for PICC line tip location | ||
| Patch-based AI approach | Fully convolutional neural network | |
| Absolute distance from ground truth: mean ± standard deviation | 4.66 ± 2.8 mm | 3.10 ± 2.03 mm |
| Absolute distance from ground truth: root-mean-square error (RMSE) | 5.44 mm | 3.71 mm |
| Algorithm execution time | 87.67 seconds per image on a GPU | 1.32 seconds per image on a GPU |
This represented 1.5 times better performance and more than 66 times faster execution on a graphics processing unit (GPU), Lee said.
In addition to its base functions of showing the trajectory of the PICC and locating the line tip, the model could also be used to spot cases with nonexistent or partially visualized PICC lines, he said. It can also be used to identify ECG cables and electrodes, for example.
"This system can be further generalized to detect the other types of lines and tubes frequently used in the clinical setting," he said. This would be a "real help for faster preliminary interpretation of triage for [incorrectly positioned] devices."
Indeed, the group plans to extend the platform to detect other types of vascular access and therapeutic support devices, according to Lee.