The algorithms that drive endoluminal navigation of the colon using CT data are not simple creations. They are responsible for complex series of computations taken in multiple steps on vast amounts of CT data. But because these algorithms form the core of both 3D colon viewing software and CAD systems, they are often a high priority for system developers.
The navigation systems are tasked with reliably finding the true centerline of the colon, a job compounded not only by the huge volume of data contained in a single dataset, but by the tortuous morphology of the colon itself. And if the software does not perform the task quickly -- optimally in a minute or less -- it risks stalling the workflow in a high-volume virtual colonoscopy center.
In a new study, Robert Sadleir, and Paul Whelan, Ph.D., from the Vision Systems Group at Dublin City University in Ireland discuss how they addressed the problem of computational overload. In a nutshell, they tweaked an earlier centerline navigation algorithm based on topological data thinning, and made it work faster and better with the addition of an optimization technique based on a lookup table (LUT).
"Ideally, a centerline calculation algorithm should generate an accurate approximation of the central path through the colon with minimal operator intervention in a reasonable amount of time," wrote Sadleir and Whelan in Computerized Medical Imaging and Graphics (June 2005, Vol. 29:4, pp. 251-258).
Early centerline algorithms were based on a technique known as onion peeling or topological thinning, which are accurate but time-consuming and highly inefficient, they explained. As a result, many recently published approaches describe more efficient alternatives to topological thinning, or modifications to the standard topological thinning protocol.
An excellent example of such a protocol was created by Ge et al, whose multistage algorithm first identifies voxels representing the colon lumen (segmentation process) using a 3D region growing algorithm, Sadler and Whelan wrote. At this point a topological thinning algorithm is used to create a skeletal representation of the segmented colonic lumen, which is further reduced to a single centered path between two end points. Surface voxel tracking and dataset subsampling are used to improve performance. The group's centerline tracking time of about 60 seconds has been shortened further in later studies, however (Journal of Computer Assisted Tomography, September-October 1999, Vol. 23:5, pp. 786-794).
Sadleir and Whelan began with a centerline calculation algorithm similar to that of Ge et al, and improved its performance by means of a LUT-based optimization technique, finally comparing the performance of both systems in a kind of time trial.
Before their virtual colonoscopy examinations, all subjects followed a low-residue diet for 40 hours followed by clear liquids for 48 hours, and then a laxative regimen. Room-air insufflation of the colon was performed to patient tolerance, and images were acquired in a single breath-hold on a four-detector scanner at 120 kVp, 2.5-mm collimation, 3-mm slice thickness, and1.5-mm reconstruction intervals, the team wrote. (The VC data were courtesy of Dr. Helen Fenlon and Dr. John Bruzzi from Mater Misericordie Hospital in Dublin.)
First the colon lumen was extracted from the dataset using a standard method in which a binary technique separates colon voxels from background voxels. The end points use a technique based on distance fields, and any background voxels found inside the lumen must be removed. Topological thinning is used to acquire the skeleton of the segmented colon.
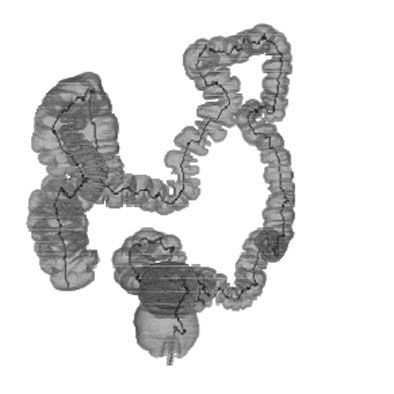 |
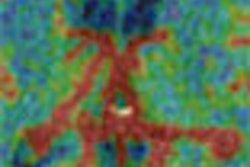
| Colon centerline calculation in which segmented data yielded a single hollow tube connecting the rectum to the cecum. Image courtesy of Robert Sadleir. |
"The thinning process involves the removal of layers of surface voxels from a binary object," the authors wrote. "A surface voxel, i.e., one that is not completely surrounded by other object voxels, is only removed if it does not affect certain structural properties of the object being thinned. These properties describe the object in terms of overall connectivity, holes, and end points. Layers of surface voxels are removed iteratively and the process terminates when no more surface voxels can be removed without compromising the structural properties outlined above. The resulting set of voxels represents the skeleton of a particular surface voxel."
Three rules are required before eluting a particular surface vowel, including end point retention, which ensures that the number and location of end points in the skeleton are the same as in the original subject. Connectivity preservation ensures the same number of distinct binary objects in a scene before and after application of the topological thinning algorithm. Third, the hole prevention rule ensures that a surface voxel is not removed if such action would introduce a new hole into the object being thinned.
However, the skeleton resulting from topological thinning usually contains extraneous loops due to holes in the segmented colon lumen that are usually associated with haustral folds. To correct them, a distance field representing the distance of the centerline from the colon surface is used to generate a weighted graph where the centerline can be identified as the "minimum cost path between two predefined end points," the researchers wrote. The Dijkstra's shortest path algorithm is an efficient way to accomplish this, they stated. Much of this process is fairly standardized, however, the authors noted.
Optimization
"Our enhancements deal specifically with the skeleton-generation phase, particularly the tests for connectivity preservation and hole prevention," Sadleir and Whelan wrote. "Both of these tests must be performed repeatedly, at least once for each object voxel, and represent the most time-consuming aspect of the centerline calculation process. In each case the 3 x 3 x 3 neighborhood surrounding the voxel under examination is extensively analyzed in order to establish whether its removal will affect the relevant global properties of the object, i.e., overall connectivity and number of holes."
The results of this analysis depend on the values of the 26 neighbors surrounding the voxel being examined. Each voxel can belong to either the object or background classes, resulting in 67,108,864 (226) neighborhood combinations.
There goes the neighborhood
The researchers were able to streamline the process by generating all possible neighborhood configurations, testing each neighborhood for connectivity preservation and hole prevention. The results were then combined into a single pass/fail binary value and stored in an LUT structure, they wrote.
"Each result is addressed by a unique index, I, that is obtained by multiplying the value of each voxel in the neighborhood (V0-V25) with a weight in the range 20-225 and then summing the results," they wrote, adding that "this is essentially a 3D convolution operation where the value of the central voxel is not used in the calculation."
Thus, the LUT process avoids the intensive computation that would otherwise be required during the thinning phase of the centerline calculation algorithm to preserve connectivity and prevent holes, they wrote. Instead, the relevant index is quickly generated and the skeleton reduced using the equation described in the previous paragraph. It is calculated only once and kept on a disk to be reloaded each time it is needed for a centerline calculation.
The researchers tested the system with and without the LUT on 12 VC datasets (six prone and six supine), all representing intact colons.
All of the resulting centerlines were perfectly formed using both systems, they wrote. But the difference in calculation time was enormous. The average calculation time using the standard approach was 155.413 seconds, compared to 3.438 seconds for the optimized LUT approach. And while it did take an hour and a half to perform a one-time calculation of the LUT data, after that the table could be loaded up for each centerline calculation in less than a second, the team reported.
As for limitations, the group cautioned that their study was applied in intact colons. In colons with collapsed segments and extreme blockages, the segmentation process can generate multiple subsections representing the air-filled colonic lumen. These subsections must be put together before the optimized centerline generation process can be activated. However, this shortcoming falls into the realm of segmentation and not centerline generation.
The software was implemented using the Java 2 standard edition version 1.4.2 (Sun Microsystems, Santa Clara, CA), and tested on a standard PC workstation running at 1.6 GHz CPU and 512 Mb RAM with Microsoft Windows XP Professional (Microsoft, Redmond, WA).
Not only did the use of an LUT relieve demand on the CPU, it made centerline calculations approximately 45 times faster than with conventional methods, the team concluded.
"Our modifications are relatively straightforward to implement and do not alter the core functionality of the topological thinning algorithm, hence the quality of the resulting centerline is not compromised," Sadleir and Whelan wrote.
By Eric Barnes
AuntMinnie.com staff writer
June 8, 2005
Related Reading
CAD struggles through tagged, subtracted VC data, May 18, 2005
Support vector machines boost accuracy of VC CAD, May 3, 2005
New VC CAD applications show promise as second reader, March 4, 2005
Iodine tagging regimen yields best VC results, January 27, 2005
Low-prep VC study finds CAD can be fooled, December 23, 2004
Copyright © 2005 AuntMinnie.com