Do you want to develop deep-learning algorithms for medical imaging applications at your institution? There are some important practical considerations to be aware of, according to an August 17 webinar sponsored by the Society for Imaging Informatics in Medicine (SIIM).
For example, you'll need data to train the algorithms and tools to curate and manage the data. You'll also need to overcome the significant barriers to conducting this type of research and then, once the algorithms have been developed and validated, consider how to apply them clinically, according to speakers during the roundtable discussion.
Getting data
 Dr. Eliot Siegel from the University of Maryland.
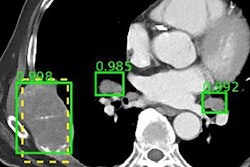
Dr. Eliot Siegel from the University of Maryland.Training deep-learning algorithms requires access to data. Fortunately, there are a variety of public databases available for use in training artificial intelligence (AI) algorithms, including the U.S. National Institutes of Health's (NIH) Cancer Imaging Archive (TCIA). The TCIA has 62 collections of data, including 4D lung images, breast MRI, CT colonography, CT of lymph nodes, etc. The collections also include the Lung Image Database Consortium (LIDC), which features lung nodules that are outlined and delineated on CT images, and the Reference Image Database to Evaluate Response (RIDER), which looks at changes over time for lung nodules on thoracic CT, said Dr. Eliot Siegel from the University of Maryland School of Medicine.
"[In addition], the [American College of Radiology] has sponsored a large number of imaging trials, and they also make their data available upon request," Siegel said.
A white paper published online May 17 in the Journal of Digital Imaging by Dr. Marc Kohli, Dr. Ronald Summers, PhD, and Dr. Raymond Geis discussed the issues involved with medical image data and datasets in the era of machine learning. The white-paper authors noted that virtually all available imaging databases use processed images instead of raw images including the k-space data for MRI and CT sinogram data, Siegel said.
"It may be that those raw data collections have tremendous value for machine learning," Siegel said. "In general, we're just looking [right now] at processed images that are optimized for the human visual system."
How many cases do you need?
Limited research has been performed on the sample size needed to train an algorithm, although there is a rule of 10 -- i.e., the number of cases needed would be 10 times the number of parameters in the algorithm. That number sounds fairly low, however, Siegel said.
"Most of the publicly available datasets tend to include tens to hundreds of cases, with very few as large as 1,000 or more," he said.
Siegel also noted that the NIH requires applicants and grantees to submit a plan to share their final research data before providing access to their databases.
"Despite that, there are many issues related to principal investigator embargoes on releasing data for some period of time and a requirement for committee approval to download data," Siegel said.
Data may be more accessible in other countries, such as Israel, China, and India, he noted.
Large institutions with medical images have traditionally been able to mine data within their own firewall, giving them a substantial advantage for developing deep-learning algorithms.
"That's why it would be great if we could have an index of publicly available and publicly accessible data to even the playing field a little bit," Siegel said.
There's also tremendous potential for large national groups such as teleradiology providers to be able to share their data in a deidentified way, he said.
Chicken or the egg?
Which comes first, the data or the question you'd like to address with machine learning? This is kind of a chicken or the egg question; both are essential, said Dr. Bradley Erickson, PhD, from the Mayo Clinic in Rochester, MN.
 Dr. Bradley Erickson, PhD, from the Mayo Clinic.
Dr. Bradley Erickson, PhD, from the Mayo Clinic."If you pick an area that you're interested in, you're probably going to be a lot more motivated to pursue the nuances and understand the clinical significance and continue to persist when the inevitable challenges arise," Erickson said. "So maybe the question comes first, but clearly you need to have datasets that are available in that area, and it may force you to adjust your focus a little bit depending on the data that's available."
Then you need to look at the data to see if there are interesting questions that can be addressed by machine learning, Erickson said.
"So you need to look at what's unique about the data, what's unique about the population, [the] longitudinal information, [or] maybe some sort of intervention that's unique, or perhaps the imaging device," he said.
Data curation is critical, and you'll need the right tools for the job if you want to pursue a deep-learning project at your institution. This includes a query application for searching radiology reports to find a dataset to train the algorithms, according to Dr. George Shih of Weill Cornell Medical Center. Once you have a list of exams you'd like to use, the exams can then be deidentified and imported, for example, into a folder on a research PACS.
"Since most of the deep-learning algorithms that are being used today are supervised learning techniques, we'll need to provide annotations for the dataset," he said. "Usually that means drawing circles around areas of interest on the images, and it's really critical to be able to do this accurately and efficiently to build the best learning algorithms. You can't really overstate the importance of the annotations."
Commercial software is available for annotation, but there are also some open-source applications such as Horos or OsiriX that can be used, Shih said. Some researchers also utilize custom software developed using the Python or matrix laboratory (MATLAB) programming languages.
Garbage in, garbage out
Data curation is needed to avoid the problem of garbage in, garbage out, so you really need good tools to efficiently identify and refine the set of exams you want to work on, Erickson said.
"You also need to have some degree of integration with your analysis, because rarely will the first pass of curation be the only pass through," Erickson said. "Sometimes as you do the analysis you realize ... that there may be some images that have motion artifact that are throwing things off. There may be errors in your pipeline analysis or your assumptions about the data. Or you may just find that you don't have enough data to get results, and you have to go back and curate some more."
It's critical that your data management approach be scalable and flexible, Erickson said.
"Rarely is the first hypothesis or question the actual or final one that you end up with," he said. "So you often go back and iterate the question that you want to ask, and that may require further curation or an adjustment of your curation process."
Traditionally, developing algorithms in medical imaging required the purchase of commercial software. Now, however, most of the libraries required for deep learning are open-source, making it easier for anyone to get started, Shih said. To develop algorithms, Shih uses a Docker container -- a development environment that he utilized to build a deep-learning framework. Algorithm training is then performed on a local computer designed for deep learning and equipped with four graphics processing units (GPUs).
Overcoming barriers
The biggest barriers to conducting this type of research include the limited access to data outside of publicly available datasets, even within one's own institution, Siegel said. It can also be extraordinarily difficult to recruit experienced data scientists with experience in deep learning, especially for medical imaging.
"There really is not a machine-learning or deep-learning undergraduate or graduate course that's available now," Siegel said. "So to a large extent we all end up finding folks who have experience in computer science or statistics or databases, etc. I'm hoping that will change as time goes on, but to a large extent a lot of these folks who are really excellent are being recruited from one place to another place, and it's hard to have a pipeline for new folks."
Also, many facilities that do not have undergraduate campus or substantial computational resources may be faced with limited resources to conduct AI research, Siegel said.
"Also, GPUs can be difficult to find and expensive in the era of people using them for video games and for bitcoin mining," Siegel said.
Siegel also noted that radiologists may have limited academic time to annotate images and to get deeply involved with deep learning. What's more, grant funding has not traditionally funded deep-learning algorithm development for imaging; it's seen as more of an engineering issue than a "new science" because deep learning has been around a long time, Siegel said.
HIPAA concerns over the risk of patient identification can make it harder to collaborate with other institutions, he noted. In addition, it can be difficult to be entrepreneurial in light of perceived regulatory barriers for commercializing these algorithms.
Clinical workflow
Once the algorithms have been developed and validated, it's important to consider how they would be integrated into the clinical workflow. One way to do it is by incorporating the algorithms into the reporting workflow, Shih said.
"We are working on an open-source framework that we will be presenting at RSNA [2017] that will allow for report generation from radiology templates," he said. "I know a lot of people are working on something similar, but I think this will allow for easy integration of machine-learning algorithms."
Most AI applications currently rely on server-based operation, but there will also be some AI cases that will require the ability to be run on local systems -- such as PACS workstations or even mobile devices, Shih said.
"An example that a lot of people give is a self-driving car, in which you have a lot of data coming to the car from the sensors and you can't send it back to the server for inferencing," Shih said. "There will be a lot of scenarios in healthcare where local inferencing will make a lot of sense because you have different pieces of data. Obviously the [electronic health record] data is stored on servers, but if you are working with a point-of-care device such as a portable ultrasound unit or your smartphone, you will need [the algorithm] to be able to inference locally."
Siegel noted that the current model for mammography computer-aided detection (CAD) uses a second-reader paradigm, in which the CAD software is applied as a second reader after the radiologist has initially reviewed the mammogram. It would be better, though, if machine-learning algorithms could be applied interactively -- highlighting areas on the image that the radiologist is looking at and providing its level of confidence and the reasons for the identification, analysis, and recommendation, Siegel said.
"Right now, if you want to consume machine-learning types of applications in general the model has been to take my dataset and send it out, have some analysis done, and then bring it back, and I can look at that as if it were a second window or second opinion," he said. "But integrating that into workflow is going to be really critical as time goes on to the success of this. A lot of what we want is improved productivity, and we want it to essentially flow in our own workflow, and that's going to require a fair amount of additional work."