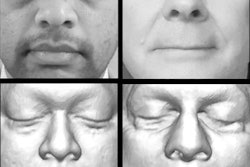
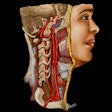
3D surface reconstructions can produce beautiful renderings of a patient's face. So beautiful that individuals could potentially, although not easily, be identified just from their reconstructed CT scan, according to a study published in the June issue of the American Journal of Roentgenology.
A team of researchers, led by first author Dr. Joseph Jen-Sho Chen from the University of Maryland School of Medicine, asked volunteers to match 3D CT surface reconstructions with one of five photographs. Overall observer accuracy was only 61%; however, the reviewers correctly identified patients in 88% of the cases in which one of the photographs matched the 3D image.
"Despite the fact that it is a difficult task, identification of a patient could definitely be made solely based on a 3D reconstructed image," senior author Dr. Eliot Siegel told AuntMinnie.com. "Our results support the idea that it is not possible to truly 'deidentify' a CT image of the face -- and probably MRI as well -- when used for teaching, research, or clinical purposes."
A HIPAA violation?
The idea for the study came from a presentation given by a lawyer at the U.S. National Cancer Institute, Siegel explained. In his talk, the lawyer suggested that they could not archive images of the head that included the neck because it was a HIPAA violation. He also said that a surface reconstruction would reveal the patient's face and that this was protected health information (PHI), Siegel said.
"My initial response was to search the literature to find out whether anyone had ever tried to determine the success rate of identifying a patient's identity from those '3D surface reconstructions,' and when I found out this hadn't been done, it seemed like an important and interesting project," he said.
 Dr. Eliot Siegel from the University of Maryland.
Dr. Eliot Siegel from the University of Maryland.
The study included 29 patients who had received clinically indicated CT scans of the maxillofacial sinuses or cerebral vasculature, which were reconstructed using a 3D workstation (AquariusNet server, TeraRecon). These patients were also photographed, along with 150 other volunteer patients (AJR, June 2014, Vol. 202:6, pp. 1267-1271).
Chen and colleagues then recruited 149 observers and asked them to match the surface-reconstructed images from the CT data with five randomized photos. Of the 149 observers, 29 (19.5%) were radiology residents, eight (5.4%) were radiology fellows, 15 (10.1%) were radiology attending physicians, 53 (35.6%) were other healthcare professionals, and 44 (29.5%) were in nonhealthcare professions.
The image reviewers were presented with a set of 58 questions in a Web-based format. For each question, they were asked to match a surface-reconstructed 3D image of a patient with one of the five digital photographs, or to choose "none of the above."
Each of the 29 3D-rendered images was used in two different questions. One question included a correct match among the five photograph choices, while the second question did not include a correct photo match.
Overall accuracy -- correctly identifying a matching digital photograph or selecting "none of the above" -- was just 61%.
"This was despite the fact that in most of the cases ethnicity of the [photo] choices and sex varied," Siegel said. "Even with those seemingly obvious cues, the success rate was still only about 3 out of 5, not much better than a coin flip."
However, sensitivity -- correctly matching a photograph with the reconstructed image -- was 88%. Specificity -- correctly choosing "none of the above" when the reconstructed image did not match any randomly displayed photos -- was only 50%. Siegel noted that the "none of the above" option seemed to make the matching task much more difficult.
"Surprisingly, only 50% of respondents were right when 'none of the above' was the correct response, while they were much better [88%] when one of the patient photographs actually did match the surface-rendered CT image," he said. "Perhaps not surprisingly, the best performances were from those observers who were the same race as the 3D CT image."
The researchers did not find any statistically significant difference, though, regarding the observers' accuracy and their sex, age, or ethnicity.
"Our research findings could be interpreted as simultaneously supporting the suggestion that remarkably lifelike surface-reconstructed images can be successfully matched with patient photographs but also suggesting that this task of identification can be quite difficult without familiar cues such as hair, skin color, and markings, and the differences in the patient's face when in the supine position for CT," the authors concluded.
Overall, the results "suggest that a form of encryption for these images should be performed even when the patient information is hidden when there is the possibility that the images could be seen or intercepted by those who do not have permission to access this PHI," Siegel said.
Other work
Siegel also referred to a follow-up study by the research team that was published in the Journal of Digital Imaging (June 2012, Vol. 25:3, pp. 347-351). In that study, the group found that Google's facial recognition algorithm (part of its Picasa 3.6 software) produced only 28% overall accuracy for identifying patients based on their 3D surface-reconstructed facial images.
"Since then, there have been major advances in facial recognition, including software recently purchased by Facebook, [called] DeepFace, which claims an accuracy of 97.5% for facial recognition," he said.
The researchers have also presented and intend to publish results from a project that created software that "masks" the face without any loss of pixels from the original image.
"This effectively prevents identification of a patient when using 3D surface-rendering software," Siegel said.