Researchers at Cincinnati Children's Hospital Medical Center (CCHMC) have developed and validated an abdominal organ segmentation model and made it available as an open-source application for further research and potential clinical deployment in pediatrics, according to an article published in the American Journal of Roentgenology on May 1.
After testing a number of different deep-learning algorithms, first author Elanchezhian Somasundaram, PhD, from the departments of radiology and pediatrics at CCHMC, reported that transfer-learning models -- algorithms trained on heterogeneous public datasets and then fine-tuned using institutional pediatric data -- yielded the best overall performance on internal and external validation.
The findings are important because "children differ from adults both in abdominal organ size and in the quantity of intraabdominal fat separating organs," authors noted. "The effectiveness of automatic deep-learning models in segmenting the pediatric pancreas is of particular concern given this organ’s small size and variable morphology."
The CCHMC team developed and tested their model to perform liver, spleen, and pancreas segmentation using 1,731 pediatric CT examinations. The exams included varying scanner types, acquisition parameters, and reconstruction techniques. All examinations were reconstructed in the axial plane. Of the 1,731 exams, 1,504 were used for training and 221 were set aside for testing.
Three deep-learning model architectures, SegResNet, DynUNet, and SwinUNETR from the Medical Open Network for AI (MONAI) framework, were trained using native training, relying solely on institutional datasets, and also transfer learning, incorporating pretraining on public datasets. TotalSegmentator (TS), a publicly available segmentation model, was applied to test data without further training. Segmentation performance was evaluated using mean Dice similarity coefficient (DSC), with manual segmentations as reference, and volume similarity (VS).
Two board-certified pediatric abdominal radiologists (with 14 and 11 years of postfellowship experience, respectively) reviewed the prior manual segmentations, accessed through an institutional shared drive, and the reviewers jointly assessed the segmentations and potentially edited segmentations after discussion.
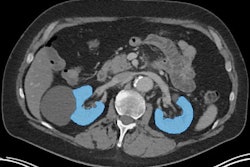
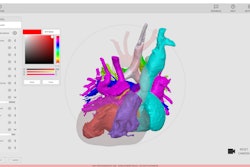
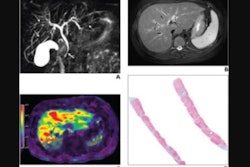
 This image shows ground-truth manual organ segmentations (yellow) vs. DynUNet-TL (green) and TotalSegmentator (red) predictions. Photo courtesy of the American Journal of Roentgenology.
This image shows ground-truth manual organ segmentations (yellow) vs. DynUNet-TL (green) and TotalSegmentator (red) predictions. Photo courtesy of the American Journal of Roentgenology.
The CCHMC team concluded that transfer learning models trained on heterogeneous public datasets and fine-tuned using institutional pediatric data outperformed internal native training models and TotalSegmentator across internal and external pediatric test data. The group added that segmentation performance was better in the liver and spleen than in the pancreas and that the algorithm will be useful for volumetry applications in pediatric imaging.
If successful, the deep-learning algorithm could ease the burdens of time-consuming manual and semi-automated methods for medical image segmentation that are difficult to accommodate into routine clinical workflows, the authors noted.
"This quantitative information when combined with organ nomograms (which we have published in earlier work) can become a power diagnosis tool for early diagnosis of abdominal disease, including possible opportunistic screening, and other downstream applications that require organ contours such as radiomics based diagnosis or patient specific organ dosimetry," Somasundaram told AuntMinnie.com in an email.
See the datasets used, training strategy, hyperparameter tuning, and processing times in the full article.