An artificial intelligence (AI) algorithm can reliably classify benign and malignant bone lesions on F-18 sodium fluoride (NaF) PET/CT images, showing promise for automating a time-consuming and subjective process for nuclear medicine physicians, according to a research team from the University of Wisconsin in Madison.
After training and incorporating a machine-learning algorithm into internally developed software for tracking bone metastases on F-18 NaF PET/CT, researchers led by doctoral student Timothy Perk found that their method could produce high sensitivity and specificity for classifying benign and malignant lesions. The algorithm's classification performance also exceeded other methods previously reported in the literature.
What's more, the algorithm could be trained to replicate the performance of individual nuclear medicine physicians, facilitating time savings for physicians and more consistent results, according to the researchers. They shared their findings at last month's American Association of Physicists in Medicine (AAPM) annual meeting in Denver.
Frequent false positives
F-18 NaF PET/CT imaging has been shown to be the most sensitive method for imaging bone metastases. However, the modality suffers from false positives, as uptake can also occur in benign disease such as osteoarthritis, Perk said. In addition, patients can have hundreds of lesions to be evaluated.
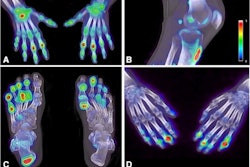
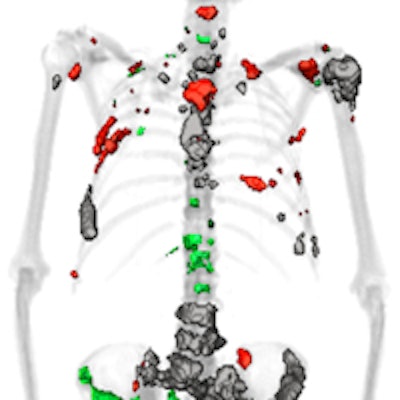
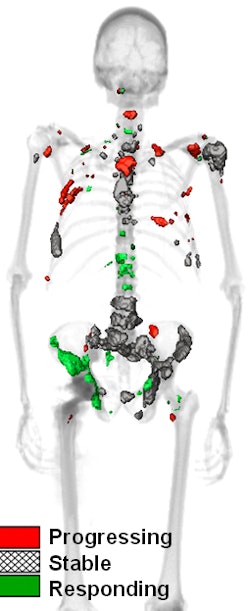 QTBI software identifies lesions that show increasing, decreasing, or stable uptake over the course of treatment. All images courtesy of Timothy Perk.
QTBI software identifies lesions that show increasing, decreasing, or stable uptake over the course of treatment. All images courtesy of Timothy Perk."We need a consistent method for whole-skeleton automated lesion classification," Perk said.
The University of Wisconsin research group has been developing software called Quantitative Total Bone Imaging (QTBI) for assessing F-18 NaF PET/CT images from patients with multiple bone metastases -- a necessary tool for tracking response in patients with hundreds of lesions, Perk said.
The QTBI software begins by automatically detecting thresholds that vary for each bone; these variable thresholds are optimized to maximize detection of disease uptake while excluding background information, Perk said. This process generates regions of interest that represent different lesions. Next, the software extracts 170 different imaging features -- including PET and CT texture and spatial probability features from the PET and corresponding CT images -- as well as a label based on the location of the lesion and information from population disease distributions.
The researchers then sought to use machine learning to also provide automated characterization of bone lesions. To train a machine-learning model, a nuclear medicine physician analyzed F-18 NaF PET/CT images from 37 bone metastatic castrate-resistant prostate cancer patients and manually identified and classified each lesion as 1 (definite metastasis), 2 (likely metastasis), 3 (equivocal), 4 (likely benign), or 5 (definite benign). One physician classified all 1,752 lesions from the 37 patients, while three other physicians also assessed a subset of 598 lesions from 14 patients. The researchers noted that the physicians agreed only 63% of the time.
After correlated features from the initial analysis by the software were removed, the final selected features were input into the machine-learning algorithm, which was based on a random-forest model and trained using tenfold cross validation. The researchers then trained and compared different models by varying the lesion detection method used (a standardized uptake value > 10 g/mL threshold; a global standardized uptake value > 15 g/mL threshold; or bone-by-bone optimized variable thresholds); the feature sets used (CT, PET, and population distribution information); and training and predicting the classifications of different physicians.
Perk and colleagues performed receiver operating characteristic (ROC) analysis to calculate the algorithm's area under the curve (AUC), and they also gathered sensitivity, specificity, positive predictive value, and negative predictive value statistics.
High performance
The inclusion of optimized bone-specific thresholds enabled the software to achieve higher classification performance than other thresholds previously reported in the literature, according to the authors.
| Classification performance | |||
| Standardized uptake value (SUV) > 15 g/mL threshold | SUV > 10 g/mL threshold | Machine-learning algorithm | |
| Area under the curve | 0.86 | 0.87 | 0.95 |
The improvement in performance was statistically significant (p < 0.0001). The model also yielded 88% sensitivity, 88% specificity, 83% positive predictive value, and 92% negative predictive value.
When trained using the classifications of different physicians, the model could also accurately reproduce how each individual classified benign and malignant bone lesions.
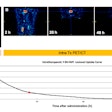
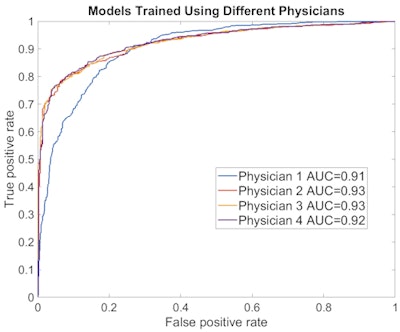 ROC curves show the model's performance when trained and tested on different physicians. The model yielded high performance for each of the four physicians in the study.
ROC curves show the model's performance when trained and tested on different physicians. The model yielded high performance for each of the four physicians in the study.As a result, the model could therefore be adjusted to predict how physicians in different clinics will classify the lesions, and it could enable efficient and consistent automated lesion classification, according to the researchers.
"Furthermore, training our machine learning on multiple physicians [can allow us to] develop optimal classification surpassing each individual physician," Perk said.
The researchers are now incorporating the machine-learning algorithm into the QTBI software, which will be submitted for approval to the U.S. Food and Drug Administration (FDA) with a corporate partner, Perk said.