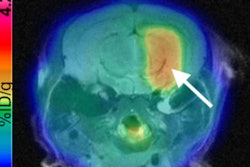
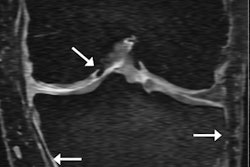
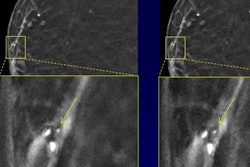
The combination of machine learning and radiomics can predict the molecular subtypes of medulloblastoma, paving the way for early diagnosis and better treatment of this common cancerous pediatric brain tumor, according to research presented last week at the American Medical Informatics Association (AMIA) meeting in San Francisco.
Researchers from Stanford University and Harvard University have trained and tested a machine-learning model that was able to predict medulloblastoma subtypes based on image textures extracted from brain MR images. By identifying these subtypes sooner, clinicians could then quickly decide on the therapeutic approach that's best for the particular subtype, according to the authors.
"MRI-based radiomic features are very predictive for molecularly defined subtypes of medulloblastoma," said presenter Mu Zhou, PhD, from the Stanford Center for Biomedical Informatics Research.
Radiomics
Radiomics -- quantitative analysis of textural image features -- has recently shown promise for enabling predictive machine-learning models to "mine" information on imaging studies to support clinical decision-making. To be clinically useful, though, radiomic properties must be clearly linked to meaningful biological characteristics, Zhou said.
 Mu Zhou, PhD, from Stanford.
Mu Zhou, PhD, from Stanford.As a result, the researchers sought to develop a machine-learning algorithm to utilize MRI-based radiomic features for predicting the four distinct molecular subtypes in medulloblastoma -- the most common malignant childhood brain tumor.
"We would like to find out the use for predictive qualities of MR imaging data to help our radiologists make better, fast, and accurate [decisions that have] therapy implications for patients," Zhou said.
Each of the medulloblastoma subtypes is associated with different clinical outcomes. The wingless (WNT) subtype offers a very good prognosis, while the sonic hedgehog (SHH) subtype has a good prognosis in infants and an intermediate prognosis in others. Patients with the group 3 subtype have a poor prognosis, however, and those with the group 4 subtype have an intermediate prognosis.
Understanding the relationship between imaging and these molecular subtypes would enable rapid assessment of the underlying cancer biology of individual pediatric brain tumors, according to Zhou.
Pilot study
In this pilot study, the researchers used a medulloblastoma database of 89 patients from Lucile Packard Children's Hospital in Palo Alto, CA; the Hospital for Sick Children in Toronto; and Boston Children's Hospital in Boston. The patients had received preoperative T1- and T2-weighted MRI. The database included subjects with all four molecular subtypes: WNT (17 cases), SHH (24 cases), group 3 (18 cases), and group 4 (30 cases). The subtypes were identified based on gene-expression profiles from molecular analysis of tissue samples.
After three board-certified radiologists manually annotated the tumor boundaries, 590 MRI-based radiomic features were extracted from both the T1- and T2-weighted imaging data. These features included the peak position of maximal intensity, entropy of histograms, histogram bin features, histogram statistical features, local area integral invariant features, tumor edge sharpness features, Daube features on the histogram, and Gabor wavelet features.
The researchers then trained a machine-learning model to predict the medulloblastoma subtypes based on these radiomic features. After performing 10-fold cross-validation to identify the optimal parameters and the number of features for the model, Zhou and colleagues found that it achieved a promising level of performance for identifying medulloblastoma subtypes, as determined by the area under the curve (AUC) from receiver operating characteristic analysis. The best results were achieved when the algorithm included radiomic features from both T1- and T2-weighted imaging data:
- AUC for SHH: 0.79
- AUC for WNT: 0.45
- AUC for group 3: 0.70
- AUC for group 4: 0.83
Clinical use?
To get a sense of how the algorithm would perform in clinical use at different institutions, the researchers decided to also train the model using datasets from just two of the three participating hospitals in the study. They then tested it on the dataset from the third hospital. This process was repeated using different sets of training and testing data from the individual hospitals, enabling the model to be tested separately on all three datasets. The researchers then calculated an average AUC from all three tests:
- Mean AUC for SHH: 0.73 ± 0.03
- Mean AUC for WNT: 0.72 ± 0.17
- Mean AUC for group 3: 0.57 ± 0.05
- Mean AUC for group 4: 0.80 ± 0.14
The experimental results showed that the MRI-based radiomic features were predictive for distinguishing the medulloblastoma subtypes, according to the researchers. Zhou noted that tumor edge and histogram-based features were the most important associations of radiomic features with medulloblastoma subtypes.
Future plans
The researchers plan to extend their method to other imaging techniques such as diffusion MRI, Zhou said. As annotations of the tumors were performed by multiple radiologists in their study, they would also like to evaluate the effect of interreader variability on the model's results.
Furthermore, they plan to evaluate the model's performance in larger clinical cohorts, as well as investigate the usefulness of radiomic features for predicting patient survival, Zhou said.