An algorithm combining the Fibrosis-4 Index (FIB-4) and an ultrasound deep-learning model could improve diagnostic accuracy and referral management for all-cause advanced liver fibrosis, a study published April 23 in Radiology found.
Researchers led by Li-Da Chen, PhD, from the First Affiliated Hospital of Sun Yat-sen University in Guangzhou, China found that their combined sequential algorithm improved specificity by over 20% for predicting pathologically advanced liver fibrosis compared with the Fibrosis-4 Index (FIB-4) alone. Additionally, the algorithm reduced unnecessary referrals by 42% without requiring access to liver stiffness measurement.
“AI-based image analysis provides a promising approach for noninvasive assessment of liver fibrosis on ultrasound images, offering an accessible tool for primary healthcare setting,” Chen and colleagues wrote.
Noninvasive tests can be used to screen patients with chronic liver disease for advanced liver fibrosis. However, the researchers noted that the use of single tests may not be enough to provide an accurate diagnosis.
Chen and colleagues constructed sequential clinical algorithms that include an ultrasound deep-learning model. From there, they compared the ability of the algorithms in predicting advanced liver fibrosis with that of other noninvasive tests.
The retrospective study included adult patients with a history of chronic liver disease or unexplained abnormal liver function test results who underwent B-mode liver ultrasound between 2014 and 2022 at three healthcare facilities.
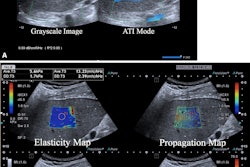
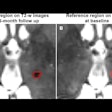
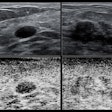
The researchers trained an ultrasound-based deep learning network (FIB-Net) on ultrasound images to predict whether the shear-wave elastography (SWE) value was 8.7 kPa or higher, which indicates advanced fibrosis. The training, validation, and test data sets included 3,067, 1,599, and 1,228 patients, respectively.
The researchers also simulated screening scenarios where liver stiffness measurements were or were not available. They did so using a two-step algorithm using the Fibrosis-4 Index (FIB-4) followed by FIB-Net in the internal and external test sets. Additionally, they used a three-step algorithm using FIB-4 followed by FIB-Net and SWE.
Finally, the team calculated diagnostic accuracy using liver biopsy as the reference standard and compared results between FIB-4, SWE, FIB-Net, and the European Association for the Study of the Liver guidelines (i.e., FIB-4 followed by SWE), along with sequential algorithms.
FIB-Net alone obtained a noninferior specificity with a margin of 5% (p < 0.001) compared with SWE (80% vs. 82%, respectively). The two-step algorithm meanwhile showed higher specificity (79%) and positive predictive value (PPV, 44%) than FIB-4 (specificity, 57%; PPV, 32%). The two-step algorithm also reduced unnecessary referrals by 42%.
Additionally, the three-step algorithm had higher specificity (94%) and PPV (73%) compared with the European Association for the Study of the Liver guidelines (specificity, 88%; PPV, 64%). It also reduced unnecessary referrals by 35%.
The study authors called for future studies to focus on validation across patients with different demographic characteristics or causes of liver disease.
In an accompanying editorial, Adarsh Ghosh, MD, from the Cincinnati Children’s Hospital Medical Center in Ohio wrote that studies such as this one show how deep learning can add value to liver ultrasound in triaging and identifying patients at high risk for fibrosis. He added that this allows for better streamlining of patient referrals and use of resources.
“Further iterations of such algorithms might also lend themselves to opportunistic screening for fibrosis in the general population undergoing liver ultrasound for other indications,” Ghosh wrote. “Future studies, however, must prioritize the development of multicenter-validated models across large population groups and disease causes for greater generalizability.”
The full study can be found here.