Researchers from Oregon have developed a novel computer-aided detection (CAD) application for radiology reports -- one that churns out differential diagnoses, accurately drawing correlations between imaging findings and potential disease states, according to a presentation at the Society for Imaging Informatics in Medicine (SIIM) meeting.
The group's "FHIRgamuts" project tested whether radiology reports can be generated automatically using data sources such as Fast Healthcare Interoperability Resources (FHIR); BioPortal Annotator from the U.S. National Center for Biomedical Ontology; and Gamuts (Radiology Gamuts Ontology), a knowledge model of differential diagnoses in radiology. They used the data to build an informatics tool populated with patients' medical histories and radiology reports to output a set of possible differential diagnoses.
"If you get rid of some of the confounding information, the differentials that you get back are actually sometimes pretty reasonable," said Dr. James Morrison, from Oregon Health and Science University, during his SIIM 2015 presentation.
A natural for radiology reports
CAD may be a natural for generating radiology reports. After all, a key job of radiologists is to synthesize patient histories, imaging findings, and physiologic data to interpret imaging exams. CAD offers a second look at the patient data to glean associations and potential disease states that may have gone unnoticed by the radiologist.
 Dr. James Morrison from Oregon Health and Science University.
Dr. James Morrison from Oregon Health and Science University."We were wondering what we could do with all of these available APIs, and we decided to do a mashup combining FHIR and Gamuts," Morrison said. "The idea was to automatically retrieve these radiology reports from the EMR, parse the text, try to identify the relevant terms, feed these terms into the Gamuts [application programming interface (API)], and see if we could get some useful results."
Among available tools, RadLex is a lexicon of standardized radiologic terms combined with ontologic associations that permit machine understanding of related anatomy, physiology, and radiologic exams. Gamuts is a collection of disease state descriptions that efficiently links symptoms, diseases, and causes. These components form the basis of computerized language processing. Add report retrieval and text processing, and the result is a CAD system that can offer supplementary differentials through the information contained in radiology reports.
Data from FHIR and BioPortal Annotator are used to interface with the RadLex pathology, which pulls up the relevant terms, Morrison said.
"We feed those terms into Gamuts, and we use the information we get back from Gamuts to try to come up with a differential diagnosis list," he said.
Morrison and colleagues Dr. Jason Hostetter, Abhi Agganwal, and Dr. Ross Filice started the FHIRgamuts project at the Hackathon at SIIM 2014. They built the JavaScript application with a Node.js framework for easy communication with APIs. A Bootstrap overlay enables HTML coding and accepts input from the HL7 FHIR interface as well as plain text. The text input is fed into the RadLex file portal annotator, which then outputs a RadLex term list that is both organized and intuitive, Morrison said.
"We take that [RadLex] term list and send it out to the Gamuts interface, and that sends us back information for each one of those terms," he said. Commonly encountered information including the important "caused by" or "may cause" descriptors are then fed into a ranking engine, which outputs a differential list.
Using the interface, if you click on radiology reports from FHIR, it outputs a list from the test database, and clicking on an individual report generates the report text. Manual entry is another option: Clicking on "Generate DDX" pulls in the report text, highlights the terms that are also present in the RadLex ontology database, and produces a differential diagnosis list.
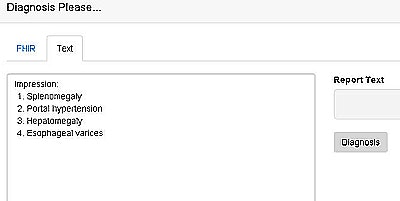 FHIRgamuts interface shows radiologist's impressions, including splenomegaly, portal hypertension, hepatomegaly, and esophageal varices. The user types in report text and clicks "Diagnosis" to generate a list of differential diagnoses. The free application is available at fhirgamuts.xrlabs.com.
FHIRgamuts interface shows radiologist's impressions, including splenomegaly, portal hypertension, hepatomegaly, and esophageal varices. The user types in report text and clicks "Diagnosis" to generate a list of differential diagnoses. The free application is available at fhirgamuts.xrlabs.com."Our initial algorithm had some surprising and somewhat irrelevant differentials," Morrison said. "One of the things we noticed is that 'lymphoma' kind of floated to the top of the list."
Lymphoma may be fairly rare, but it is associated with a lot of disease conditions -- 359, in fact, if one inputs hepatomegaly, lymphadenopathy, splenomegaly, and anemia into the Gamuts system, with the command to look for "may cause" or "may be caused by." The group is building a ranking algorithm that can dim the importance of diseases with multiple associations.
Generally speaking, if one inputs simple text, the technique generates a "pretty reasonable set of differentials; at least it's on par with what you'd expect," Morrison said. Unfortunately, inputting complex text really amps up the variability of the output, he added.
To test the accuracy of FHIRgamuts, the team developed a collection of 50 anonymized CT abdomen/pelvis reports, feeding clinical histories and the imaging findings into the application and then using the radiologists' impression from those reports as the gold standard. BioPortal Annotator was used to add more protection for personal health information, and a grading system was used to assess the accuracy of the differentials versus human impressions.
"We did it with raw reports, and after looking at the results, we decided to see what would happen if we did some manual parsing of the input to remove things like section titles -- like 'chest, abdomen, pelvis,' " he said. Negations such as "no evidence of appendicitis" were also removed, along with references to anything deemed normal.
| Accuracy of FHIRgamuts vs. human readers | ||
| Accuracy | Raw reports | Reports using parsed data |
| High | 1 | 10 |
| Medium | 1 | 21 |
| Low | 48 | 19 |
Raw reports generated fairly poor results. However, eliminating some of the confounding information resulted in "pretty reasonable" differentials, Morrison said. After parsing the data, many examples were similar to the gold standard list produced by the impression.
The method is a radiological Turing test, or a machine's ability to exhibit intelligent human behavior, he said. If there's an attending physician on one side of the wall, and the computer and a radiology resident on the other, can the attending tell the difference?
"At this point, I'd say we're not quite there yet, but this project is a pretty good example of how you can build a fairly complex system from simple components," Morrison said. "It also shows the power of combining the unique resources that are out there in an interesting way."
Limitations of FHIRgamuts include the method's dependence on its component data resources. Still, any enhancement of RadLex or Gamuts automatically enhances the diagnostic power of the application, Morrison said. Another limitation is the inherent difficulty that computers have in differentiating between positive and negative findings. For a computer, "no evidence of appendicitis" is often identical to a perforated appendix -- a problem that can be ameliorated with advance parsing of the data in which negatives and normals are removed.
Potential future uses include data mining existing radiology reports to determine how accurately the tool can suggest diagnoses, he said. It could also be used in real time to support radiologists in looking for possible diagnoses. Finally, adding the ability to mine clinical data from other sources such as EMRs could help summarize clinical information for radiologists' use.
"The opportunity exists for CAD to play a role in assisting in diagnostic interpretation, and certainly there are other projects out there," Morrison said. "Resources such as RadLex and Gamuts are very powerful and necessary components to build an engine like this. Open-source APIs have really lowered the barrier to pulling out this kind of information and using it in unique ways."