It's not easy to develop radiology artificial intelligence (AI) algorithms that don't just perform well on data that are similar to what they are trained on. But there are ways to help improve AI robustness, according to June 24 talks at the virtual annual meeting of the Society for Imaging Informatics in Medicine (SIIM).
Indeed, opportunities exist for addressing current radiology AI challenges such as poor generalizability of AI models, difficulties in leveraging data from multiple sites, low adherence to annotation standards, and the lack of tools for evaluating AI in practice, according to Dr. Daniel Rubin of Stanford University.
Rubin and Jayashree Kalpathy-Cramer, PhD, of the Massachusetts General Hospital (MGH) & Brigham and Women's Hospital (BWH) Center for Clinical Data Science discussed the challenges and opportunities in developing radiology AI models during an educational session at SIIM 2020.
Model generalizability
Most AI models are trained using data from one or a few institutions, and they may not generalize well to new data that isn't representative of the data on which the algorithm was trained, Rubin said. This can be due to biased data, as well as differences in patient populations and imaging equipment or parameters. Rare disorders or abnormalities may also be underrepresented in the training dataset, he said.
One of the ways to address this challenge is by leveraging currently available annotated data and expanding it in different ways, Rubin said. This commonly involves the use of techniques such as data augmentation (rotating or flipping some of the data) or transfer learning (repurposing a model that was previously trained on a related task as a pretraining model for a new task), he said.
"But ultimately, that isn't enough," he added. "In both of these cases, you're working with annotated data in hand, and you really need to get as much annotated data as possible. It's impossible to get an infinite amount of quality annotated data, because it's very costly to get these annotations done."
Another possibility is to make use of the vast amount of unannotated image data contained in an institution's PACS. This can be accomplished with "weak" learning, a deep-learning paradigm that enables algorithms to be trained using unannotated data such as radiology reports.
Essentially, algorithm developers write labeling functions, or programs, that process text in the radiology reports and then generate "weak" labels, such as a particular diagnosis. After a generative model "de-noises" the labels, the resulting probabilistic labels are then used to train a model.
"There is noise in these labels, but in large volume, you can actually obtain a model whose performance will be competitive with a model trained with a lot of quality labeled data," he said.
Data from multiple sites
Assembling training data from multiple sites can engender thorny technical, privacy, legal, and intellectual property issues. However, federated learning -- in which the model is brought to the data instead of the data being brought to the model -- can help to address those challenges, according to Rubin.
This approach can yield the benefits of training an AI model using data from multiple sites while avoiding the headaches and issues of sharing the patient data. The actual nuts and bolts of how federated learning is best performed in radiology are still being researched, however.
On the downside, the performance of models built using federated learning may be inferior to those trained with centralized data, Rubin said.
"There is no free lunch here," he said.
The big variable that affects the quality of the final model from federation is the heterogeneities in the medical data across the different sites, Rubin said. These include differences in patient populations, disease manifestations, scanning hardware/parameters, image resolution, and image quality.
Other challenges with federated learning include the variation in how sites label their data and the need for sufficient IT hardware and extraction of the data from the PACS at each site, he said.
Methods do exist, however, for optimizing federated learning to account, for example, for sample size and label distribution variability, he said. These can improve results.
Image annotation standards
Another big challenge for developing robust AI algorithms is the lack of standards among institutions for image annotations, which include text labels -- such as diagnoses, findings, and imaging modality -- and regions of interest on the image.
To enable interoperability between sites, standardized terminology needs to be used for image labels and standardized formats need to be applied for storing the image annotations, Rubin said.
RadLex is the standard terminology that should be used in radiology, he said. The Annotation and Image Markup (AIM) standard has emerged as the preferred approach for saving image annotations in a standardized format, Rubin said. AIM has been added to the DICOM Standard as DICOM-SR TID 1500.
"If commercial platforms universally adopt this, then there will be interoperability across platforms for annotations," he said.
In the meantime, Rubin's group at Stanford has developed ePAD, an open-source AIM-compliant image viewing and annotation application. Achieving interoperability between sites for annotations will make it easier to train machine-learning algorithms, according to Rubin.
Evaluating AI in practice
Tools need to be developed for collecting feedback for assessing the AI algorithms once they've been placed into clinical practice, Rubin said. These metrics need to be collected to be sent back to the vendors and the community to determine how well these algorithms work and how they improve care, he said.
Both the radiologist and data from the electronic medical record (EMR) may be needed to determine the ground truth of every case that's evaluated, he said. Information about whether the AI algorithm was right or wrong needs to be collected and sent to a registry, such as an AI metrics registry being set up by the American College of Radiology (ACR).
"We'd like to populate [the registry] somehow in a workflow where radiologists, when they look at images, will establish a ground truth, assess agreement or disagreement, and record it as part of their workflow in a manner that can be transmitted to this registry," he said.
Working under an ACR Innovation grant, Rubin and colleagues are developing a prototype of a toolkit for collecting AI performance metrics in the clinical workflow.
Using the ePAD platform, radiologists can annotate images, view annotations provided by the AI, and then correct any incorrect AI annotations, he said.
"That will result in an agreement or disagreement with the AI output, so that delta between what the AI said and what the radiologist corrected becomes data to be, first of all, used to compute how the AI is performing across a cohort of patients," Rubin said. "But very importantly, those deltas can be fed back to a registry, such as that maintained by the ACR, to start tracking performance in the field."
A difficult challenge
Although it's gotten easier to develop AI algorithms that perform well on the data on which they were trained, it's very difficult to create models that are broad, robust, unbiased, applicable in other populations, self-aware of their limitations, and capable of providing measures of uncertainty in their findings, according to Kalpathy-Cramer.
For example, many recent research studies describing extremely high performance for AI models in detecting COVID-19 suffer from significant dataset biases, such as using different datasets for normal cases vs. COVID cases, she said.
"[This leads] to highly biased results that are not generalizable beyond the data on which they were trained on," she said.
Building robust AI algorithms requires considering the data curation process, assessing model brittleness, building trust in AI algorithms, and thinking about how to evaluate AI in clinical practice, she said.
Data curation
The sources for ground truth data used to train algorithms depend on the task and include radiology reports, expert annotations, pathology, and outcome data, she said. For example, the goal of identifying pneumothorax on chest radiographs could be cast in multiple different ways: a classification task (does this patient have a pneumothorax); a detection task (i.e., what region is the pneumothorax in?); or a segmentation task (what are the boundaries of the pneumothorax?), according to Kalpathy-Cramer.
"By casting that as three different problems, the annotation requirements, as well as the data size requirements, can be quite significantly different, and the way in which the machine-learning algorithms are trained is also different," she said.
Image annotations can also be performed at many different levels: patient, study, image series, image slice, or voxel. Also, the number of images needed to train a model depends on the task, she said.
One challenge with the use of image annotators for the ground truth is the potential for substantial interrater variability in many tasks. As a result, the noisier the data, the greater the need for larger datasets, according to Kalpathy-Cramer.
"It is highly recommended when you're training these algorithms to have multiple raters rate each image, at least for the test set, because there's a lot of variability in some of these tasks," she said.
Brittle models
As the performance of many AI models can degrade quite severely when applied to data outside the distribution of the training data, it's important to make sure when you're building an algorithm that you're testing it on different scenarios, such as on different scanners, populations, disease prevalences, settings, and annotators, she said.
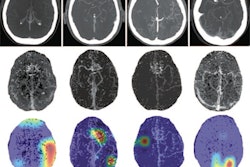
There's a great deal of interest in making deep-learning models "explainable" to users to build trust. Algorithms that provide classification results tend to be especially opaque, so post-hoc methods such as saliency maps have become popular for explaining decision-making.
However, Kalpathy-Cramer and colleagues have found variability in results from saliency maps, and she suggests that localization networks or quantitative validation be used instead.