Using a deep-learning model with lung CT imaging shows promise for identifying and segmenting tumors, researchers have reported.
The study results could have important implications for lung cancer treatment, according to lead author Mehr Kashyap, MD, of Stanford University School of Medicine. The findings were published January 21 in Radiology.
"Our study represents an important step toward automating lung tumor identification and segmentation," Kashyap said in a statement released by the RSNA. "This approach could have wide-ranging implications, including its incorporation in automated treatment planning, tumor burden quantification, treatment response assessment, and other radiomic applications."
Finding and segmenting lung tumors on CT scans is key to monitoring cancer progression, evaluating treatment responses, and planning radiation therapy, the group explained. But manual delineation is labor-intensive and subject to physician variability, it noted.
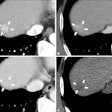
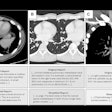
To address the problem, Kashyap and colleagues developed and tested a 3D U-Net-based, image-multiresolution ensemble deep-learning model for identifying and segmenting lung tumors on CT scans. The team trained the model using data from 1,504 CT scans and the corresponding clinical lung tumor segmentations taken from 1,295 patients treated with radiotherapy for primary or metastatic lung tumors. The researchers evaluated the model's sensitivity, specificity, false positive rate, and Dice similarity coefficient (DSC, with 1 as reference) and compared model-predicted tumor volumes to physician-predicted ones.
The group reported the following:
- The model showed 92% sensitivity and 82% specificity for detecting lung tumors.
- For a subset of 100 CT scans with a single lung tumor each, the model achieved a median model-physician Dice similarity coefficient of 0.77 and an interphysician Dice similarity coefficient of 0.8.
- Segmentation time was shorter for the model than for physicians (mean 76.6 seconds versus 166.1 to 187.7 seconds; p<0.001).
"By capturing rich interslice information, our 3D model is theoretically capable of identifying smaller lesions that 2D models may be unable to distinguish from structures such as blood vessels and airways," Kashyap said.
Kashyap and colleagues hope the study findings could help radiology departments establish their own medical image segmentation datasets.
"[Our] dataset was retrospectively assembled from routine pretreatment CT simulation scans of patients undergoing radiotherapy through a scalable pipeline involving an automated data extraction process and a custom web application for efficient clinician review of segmentation quality," they concluded. "We believe that this pipeline can serve as a blueprint for other institutions to generate their own medical image segmentation datasets."
The complete study can be found here.