A group in the U.K. has developed neural networks for interpreting chest x-rays that could help speed the adoption of AI systems in clinical settings, according to a study published December 8 in The Lancet Digital Health.
In a retrospective cohort study, the researchers developed X-Raydar-NLP (natural language processing) and X-Raydar, deep neural networks that can accurately classify 37 common chest x-ray findings from images and their free-text reports. Moreover, they have made the neural networks available to others.
“By making our model freely available, we hope to accelerate the adoption of automated systems in clinical settings to help relieve acute pressures on healthcare systems in the post-COVID-19 environment,” wrote co-first authors Yashin Dicente Cid, PhD, and PhD student Matthew Macpherson, of the University of Warwick.
AI systems for automated chest x-ray interpretation that perform at close to human expert levels are now a reality and models that are trained on large data sets can overcome generalization concerns, which have held back deployment, according to the authors.
Yet there are few freely accessible AI systems trained on large data sets for practitioners to use with their own data with the goal of accelerating deployment, they noted.
To that end, the group first compiled the largest chest x-ray data set to date in the U.K. Six hospitals from three National Health Service (NHS) Trusts contributed 2.5 million chest x-ray studies from a 13-year period (2006-2019), which yielded 1.94 million usable free-text reports written by radiologists and 1.89 million frontal x-rays.
To handle the labeling of the images, the team then developed a custom NLP algorithm (X-Raydar-NLP) to automatically extract comprehensive binary labels from the free-text reports for each image using a 37-finding taxonomy. The labeled images were then used to test their computer vision algorithm, X-Raydar, in a fully supervised machine-learning setting.
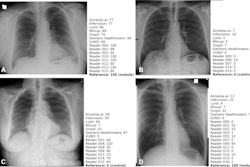
The algorithm was evaluated on three retrospective datasets: a set of exams sampled randomly from the full NHS dataset annotated by X-Raydar-NLP (n = 103,328), a consensus set sampled from all six hospitals annotated by three expert radiologists (n = 1,427), and an independent dataset named MIMIC-CXR also consisting of NLP-annotated exams (n = 252,374).
For identifying significant clinical findings, X-Raydar achieved a mean area under the curve (AUC) of 0.92 on the autolabeled set, 0.86 on the consensus set, and 0.84 on the MIMIC-CXR test.
Moreover, the algorithm’s performance on the consensus set was similar to that of the radiologist reporters for multiple clinically important findings, including pneumothorax, parenchymal opacification, and parenchymal mass or nodules, and it was noninferior on nine and inferior on one finding, the group reported.
“Our study shows that automated classification of chest x-rays under a comprehensive taxonomy can achieve performance levels similar to those of historical reporters and exhibit robust generalization to external data,” the group wrote.
The researchers have made the system available via a user-friendly web portal that allows DICOM images to be uploaded without technical barriers, with the intent of facilitating clinical implementation of AI in resource-poor healthcare systems, they noted.
“This is promising evidence towards the goal of developing a single deep-learning system that could be applied in health systems globally with confidence,” the researchers concluded.
The full study is available here.