Tracking liver metastases on follow-up CT scans is a critical task for clinicians, but the process is time-consuming and often inefficient. A new automated method for detecting and segmenting liver metastases may help, according to an article published online in the Journal of Medical Imaging.
Designed to operate as a radiologist would, the algorithm identifies, tracks, and segments lesions to assess changes between baseline CT exams and follow-up studies. In testing, the technique yielded high sensitivity for detecting metastatic liver lesions, in addition to a 90% accuracy rate for tracking tumors on a set of follow-up cases, according to biomedical engineering doctoral student Avi Ben-Cohen and colleagues from Tel Aviv University and Sheba Medical Center in Israel.
"The initial results presented here look promising," they wrote. "We achieved high sensitivity and high Dice segmentation results as well as a high matching rate. ... Using the [Response Evaluation Criteria in Solid Tumors (RECIST)] diagnosis criteria, we obtained a success rate of 88%. Additionally, expert ratings of the automated system segmentation results were in strong agreement."
An enormous task
The liver is a common site for tumor metastases from melanoma, breast cancer, colon cancer, and neuroendocrine and pancreatic cancers. Tracking, documenting, and treating them successfully represents an enormous task requiring considerable effort; usually more than one metastatic lesion is present and tumor size varies extensively. Follow-up CT is acquired every few months, with clinicians mainly looking for changes in tumor size, typically revealed by a time-consuming side-by-side comparison of two 3D CT scans, according to the researchers.
"The difficulty of this task highlights the need for computerized analysis to assist clinicians in the detection and evaluation of the size of liver metastases in follow-up examinations," Ben-Cohen and colleagues wrote (J Med Imaging, August 19, 2015).
In clinical practice, physicians compare the longest diameter for corresponding lesions on baseline and follow-up scans. Reflecting that practice, the group's algorithm relies on relatively unchanging structural information in the tissues surrounding the lesions.
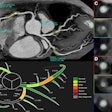
The technique employs a baseline slice method instead of using the entire exam. Then, beginning with a 2D baseline segmentation mask to identify the lesions from the first CT scan in the second one, the algorithm locates tissues surrounding the tumor using nonrigid image registration and template matching techniques, according to the researchers.
"Our algorithm imitates the radiologist's way of work by first searching for the corresponding slice in the follow-up scan, which should have a similar anatomic appearance, then searching for the lesion and finally measuring its size," they wrote.
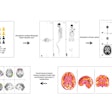
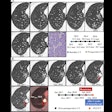
A single cross-sectional slice of a baseline CT scan acquired in the portal phase is input into the algorithm, with a lesion marked by an expert radiologist. Follow-up CT slices are then added. The analysis consists of four main stages: finding a slice containing the lesion in the follow-up scan, constraining the search for that lesion based on the prior CT, segmenting both the baseline and follow-up lesions, and, finally, propagating the 2D segmentation mask to the 3D segmentation, the group wrote.
High accuracy
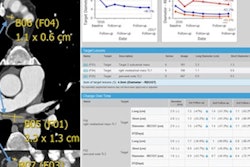
The methodology was developed using 22 cases and was tested on an additional 105 cases. The algorithm detected 94 of the 105 lesions, for an overall matching rate of 90%, and it rendered the RECIST 1.1 evaluation correctly in 88% of cases, according to the researchers.
They also tested segmentation performance using the Dice index, a statistic used to compare the similarity of two values. In this study, the Dice index average was 0.83 ± 0.08. The average sensitivity was 0.82 ± 0.13 and positive predictive value was 0.87 ± 0.11. In 92% of the cases, radiologists classified the results as acceptable or better.
The toughest challenges for identifying the correct lesion are thought to be the presence of multiple metastases, a large change in patient position between scans, and differences in contrast agents. A key strength of the method is that using the baseline CT slice instead of the entire volume "provides a prior regarding important locations," delivering slices similar to the baseline slice rather than having to search for the best alignment between two 3D datasets with no prior, the group wrote.
Better technique
Previous studies relied primarily on semiautomatic segmentation, while the algorithm developed for the current technique automatically detects the lesion at follow-up CT. All results are obtained without human intervention, Ben-Cohen and colleagues noted. The method is lesion-specific, in contrast to other methods that are designed to work on all liver lesions. The technique was modified from work by Xu et al, who designed a scheme for lymph nodes, they added.
As for limitations, the algorithm relies on the assumption that the tissues surrounding the lesion do not change much between baseline and follow-up scans, and this assumption can be incorrect if the patient has surgery between scans, if lesion size changes enough to deform the liver, or if there is another condition that changes the surrounding structures, the group wrote. If so, the registration might fail to find the correct slice and produce an incorrect segmentation.
Also, the algorithm was developed to segment lesions in 2D, and although the segmentation mask was propagated to 3D, its accuracy will require further validation. Finally, the algorithm cannot detect large morphological changes in lesions, such as merging, splitting, or disappearing, Ben-Cohen and colleagues concluded.