SAN JOSE, CA - Deep-learning algorithms based on convolutional neural networks (CNNs) are advancing a number of cherished goals in lung cancer analysis that will soon help patients live longer -- and even predict how long they might expect to live, according to a May 10 talk at the NVIDIA GPU Technology Conference (GTC).
Researchers from Stanford University discussed three of their current projects to advance key clinical goals -- all of which leverage machine learning of lung cancer features and characteristics to predict how similar features will manifest clinically. With rapid analysis of CT images, computers are even able to optimize the treatment of similar cancers in different patients.
Persistent high mortality
Lung cancer claims more than 150,000 deaths per year in the U.S. alone, and the need for new tools and techniques to help clinicians improve their decision-making is urgent. Because all lung cancer cases are diagnosed and managed with imaging, principally CT, "this is really a golden area for developing models in healthcare," said Mu Zhou, PhD, a postdoctoral fellow in biomedical informatics, who shared the stage with co-author Edward Lee, a PhD candidate in electrical engineering at Stanford.
 Mu Zhou, PhD, and Edward Lee at GTC 2017.
Mu Zhou, PhD, and Edward Lee at GTC 2017."CT is a great data source for lung tumor detection with so much information that can be quantified and analyzed in data-driven analysis," Zhou said. Toward this end, the talk highlighted three current projects underway at Stanford that seek to do the following:
- Classify the malignancy potential of lung nodules via a CNN algorithm
- Transfer the knowledge learned by radiologists in lung cancer detection to a CNN algorithm
- Predict the survival of lung cancer patients from initial CT scans
For the first project, using deep learning to better classify malignancy "will allow radiologists to stratify patient subgroups in the early stages so that personalized treatment can be used," Zhou said. "Our goal is to identify a set of discriminative features identified from CNN models."
The process involves extracting data called "nodule patches" from CT scans to capture the CT information. These patches are then fed into the CNN algorithm simultaneously to compute discriminative features, Zhou said. A total of 50 of the most discriminative CNN features are left to represent each nodule after redundant information is removed.
The study included 1,010 patients who had CT scans with markup annotations from a publicly available database. Radiologists rated each nodule for potential malignancy on a scale of 1 to 5, with 5 being the most likely to be malignant. A score of 3 was used as a cutoff between likely malignant and likely benign.
On the CNN side, a principal component analysis (PCA) sampling scheme was applied to sort features that were most likely malignant from those that were benign for each nodule. In both the training and test sets, the features showed strong performance, generating about 78% classification accuracy using both random forest and support vector machine classifiers.
"Our preliminary analysis showed that CNN models are helpful in finding nodule-sensitive information given the large range of nodule dynamics," Zhou said. "We expect with more image data available we would get further improvement."
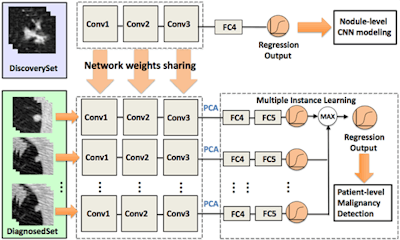 Convolutional neural networks are gaining enormous interest in cancer image diagnosis; however, insufficient numbers of pathologically proven cases impede the evaluation of CNN models at scale. The authors proposed a framework that learns deep features for patient-level lung cancer detection. The proposed model can significantly reduce demand for pathologically proven data, potentially empowering lung cancer diagnosis by leveraging multiple CT imaging datasets. Images courtesy of Mu Zhou, PhD, and Edward Lee.
Convolutional neural networks are gaining enormous interest in cancer image diagnosis; however, insufficient numbers of pathologically proven cases impede the evaluation of CNN models at scale. The authors proposed a framework that learns deep features for patient-level lung cancer detection. The proposed model can significantly reduce demand for pathologically proven data, potentially empowering lung cancer diagnosis by leveraging multiple CT imaging datasets. Images courtesy of Mu Zhou, PhD, and Edward Lee.Developing transferable CNN features
A second project at Stanford aims to develop transferable deep features for patient-level lung cancer prediction between different clinical datasets.
"The key question we would like to address is that CNN models are powerful for learning image information," Zhou said. "But currently we are facing a challenge in that we don't have large-scale clinical data of imaging plus clinical annotations," that is, pathologically proven cases. In short, a new type of database is needed that allows researchers to predict clinical outcomes without vast numbers of cases, he said.
There are two questions: Can CNN features generalize to other imaging databases? If so, how do these features transfer across different types of datasets?
The researchers are proposing the creation of a "transfer learning framework" that would transfer the knowledge acquired by radiologists to a CNN algorithm. The knowledge would come from two sets of data: one from a "discovery" dataset based on the judgment of radiologists about malignancy but without pathological confirmation. A second diagnostic dataset would contain biopsy-confirmed lung cancers or benign cases.
Testing on the diagnosed set showed that the CNN transfer data achieved 70.69% accuracy versus a slightly better 72.4% for the radiologists.
"The model captured the estimated information from input of our radiologists quite well in the discovery set," Zhou said. Not only can knowledge defined by radiologists be learned by a CNN model, the framework could potentially reduce the demand for pathologically confirmed CT data. Once again, the analysis will likely improve with the availability of more data, he said.
Predicting lung cancer mortality
Finally, can CNN data predict lung cancer survival times based on CT data alone, and, if so, can it accomplish this task better than conventional techniques?
A challenging task, to be sure, "but the reason why this is important is that we can better design specialized treatments based on better predictions of survival," said Edward Lee. "This could mean the difference between a patient going to hospice versus staying at a hospital."
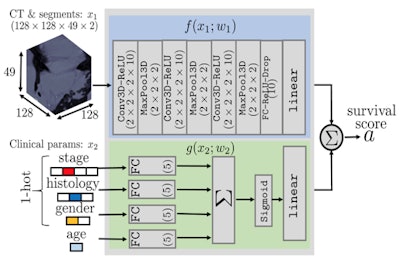 This schematic describes an algorithm to predict the survival times of each patient with lung cancer given only images and clinical data. The model uses CT data and segmentation volumes of interest in the first network and clinical information in the second network, combining both to generate a survival score.
This schematic describes an algorithm to predict the survival times of each patient with lung cancer given only images and clinical data. The model uses CT data and segmentation volumes of interest in the first network and clinical information in the second network, combining both to generate a survival score.Conventionally, a 2D CT image would be fed into a feature selection algorithm combining a range of intuitive features such as tumor density, shape, textures, and wavelets to create radiomic features, applying support vector machines or other known methods to generate a survival prediction via a Cox proportional hazards model, Lee said. Using these methods, Harvard University researchers achieved a concordance index of about 0.65 (1.0 would represent perfect accuracy) versus actual lung cancer mortality data.
For the CNN method, Lee and colleagues used four different lung cancer datasets comprising more than 1,600 patients. Three datasets had mortality information, and the researchers also included information from the patients' electronic health records such as age, sex, cancer stage, and histology.
The data were fed into a segmentation model containing predicted segmentation and 3D regions of interest, which dealt with clinical information separately from the nodule information and output a survival prediction score.
The investigators found wide variations in survival time between the cohorts, perhaps due to demographic differences not accounted for in the model, Lee said. But the CNN predictor outperformed the non-CNN model. With the CNNs alone, the group achieved 0.69 accuracy, compared with 0.65 for the Harvard group's non-CNN analysis. Moreover, when the demographic information was included with the CNN calculation, accuracy rose to 0.71.
"When we compared our model with the pretrained 2D CT model, we got very good performance," Lee said.
The researchers are currently looking at what parts of the CT image affect the survival scores, and to what degree.
"We envision a day when clinics can share models instead of data," Lee said. "This is really important because we can now bypass HIPAA, and it's computationally very efficient. The models themselves are just 1 MB, versus the storage size of 1 GB per patient if you're using DICOM images."