SAN JOSE, CA - With so much research focused on disease diagnosis using artificial intelligence, it's easy to overlook a largely untapped resource that could wield at least as much firepower to reinvent healthcare for the better. That resource is the deep patient, according to a May 11 talk at the NVIDIA GPU Technology Conference.
How do you build the deep patient? In a nutshell, you use deep learning to process patient data and derive representations of the patient that can be used to predict diseases that might develop -- and do it better than current methods, explained Riccardo Miotto, PhD, a data scientist at Icahn School of Medicine at Mount Sinai in New York City.
"The idea ... is basically to use deep learning to process patient data in the electronic health record ... that can be effectively used to predict future patient events and for other unsupervised clinical tasks," Miotto said in his talk.
Predicting outcomes from raw data
The deep-patient framework involves extracting electronic health records (EHRs) from a data warehouse and aggregating them by patient. The warehouse includes different kinds of data, such as structured data in the form of lab tests, medications, and procedures; unstructured data, including clinical notes; and demographic data such as age, gender, and race.
 Riccardo Miotto, PhD, from Icahn School of Medicine at Mount Sinai.
Riccardo Miotto, PhD, from Icahn School of Medicine at Mount Sinai.The deep-patient process takes the different data types and normalizes clinically relevant disease phenotypes, grouping similar concepts in the same clinical category to reduce information dispersion, he said. This grouping is known as a "bag of phenotypes." Users can then employ the data to make predictions about patient outcomes, perform drug targeting, predict patient similarities, or do other data modeling tasks.
For the study, Miotto led a team that modeled the data architecture on a pipeline that had worked in the lab. In the study model, the EHR data were modeled as a series of layers, with each successive layer using as input the output of the preceding layer. The last layer was the deep-patient representation.
For this, "we took the diagnosis, particularly the ICD-9 codes, we took lab tests -- not the results, just the presence of the tests -- we took medications and procedures, and we parsed the clinical notes, extracting terms using annotators, and then applied modeling on the clinical notes," Miotto said.
For every patient, the data were averaged into a single representation, and then demographic data were incorporated, averaging the phenotypes and aggregating them by patient into the aforementioned bag of phenotypes.
"We were just counting the number of times each phenotype was appearing, so basically we didn't handle time in this study," Miotto said. The data were incorporated into the network framework to create the patient representation used for analysis, which can include a wide range of tasks such as data clustering, similarity, and topology analysis.
"You can then predict events, you can use standalone classifiers, and you can fine-tune the network so it's pretty approachable for differently skilled people," he said.
Predicting disease
The study predicted the possibility that a patient would develop a new disease within a certain amount of time given his or her current clinical status. The training set included about 1.6 million patients treated between 1980 and 2013.
The test set included 100,000 patients with new diagnoses in 2014, evaluating 79 diseases based on ICD-9 codes such as endocrinology, cardiology, and oncology. Outliers were removed.
"We trained the patient model, we trained the classifier on top of the training set, we applied the model to the test data so that every patient was represented as a vector of disease risk probabilities, and we were evaluating these predictions over different time windows," Miotto said.
The results were compared with other techniques such as raw EHR, principal component analysis (PCA), k-means, and generalized method of moments (GMM) analysis -- and it outperformed them all, producing an area under the curve (AUC) of 0.79 versus the other methods.
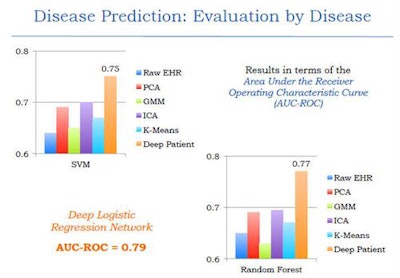 The deep-learning network outperformed other common methods of data-based disease prediction. Image courtesy of Riccardo Miotto, PhD.
The deep-learning network outperformed other common methods of data-based disease prediction. Image courtesy of Riccardo Miotto, PhD.Different classification methods including random forest classifier and support vector machines were also tested and found to be effective. An analysis of results based on disease prediction with strict time periods was also more effective than current models, Miotto said.
| Accuracy of deep-patient model for disease prediction | |
| Disease | AUC |
| Liver cancer | 0.93 |
| Regional enteritis and ulcerative colitis | 0.91 |
| Congestive heart failure | 0.90 |
| Chronic kidney disease | 0.89 |
| Personality disorders | 0.89 |
| Schizophrenia | 0.88 |
| Multiple myeloma | 0.87 |
| Coronary atherosclerosis | 0.84 |
Patient stratification
The group is working on a new project involving patient stratification in an attempt to understand the complexity of patient populations, Miotto said.
Imagine that you have a cohort of patients with a certain disease, he said. The task is to classify them, see what they might have in common, and see if they can be subgrouped into certain clusters. The patients may have common problems or similar characteristics that, once analyzed, could lead to better drugs or treatment paths.
The study used the Mount Sinai BioBank containing information on everything from DNA, RNA, and the patient's microbiome to diagnoses, drugs, and procedures employed.
"The idea is to design more precise risk models and maybe develop better drugs for patients," Miotto said.
The deep-patient model enables the leveraging of EHRs toward improved patient representation, he concluded. In terms of cons, the individual patient representations are not interpretable, and future studies need to better incorporate the element of time.
In response to an audience question seeking real-world examples of patients helped by such analyses, Miotto said it was a great question but that it comes a couple of years too early.
Another audience member said that most chronic diseases are merely a collection of symptoms based on lifestyle choices, and recommended that a more holistic approach to modeling based on this knowledge might improve the results in future studies.