With the aid of a deep-learning model, a team of researchers has detected with greater than 70% accuracy active lesions in patients with multiple sclerosis (MS), potentially helping them avoid repeat MRI scans and multiple doses of gadolinium-based contrast agents (GBCAs), according to a study published in the February issue of Radiology.
The team acquired images from some 2,000 MRI scans and used the data to train a convolutional neural network and test a connected network to detect active multiple sclerosis lesions. Their hope was to be able to use the algorithm to eliminate the need altogether to use gadolinium contrast to follow MS patients.
"The ability to identify enhancing lesions without GBCA administration by using [deep learning] can potentially minimize the need for GBCA administration, improve patient safety, and reduce costs associated with clinical care," wrote lead author Ponnada Narayana, PhD, and colleagues from the University of Texas Health Science Center and Icahn School of Medicine at Mount Sinai (Radiology, February 2020, Vol. 294:2, pp. 398-404).
Patients with MS are likely to undergo repeated, periodic MRI scans with a GBCA to evaluate hyperintense lesions. Clinicians target active lesions with T2-weighted, proton density-weighted, and fluid-attenuated inversion-recovery (FLAIR) imaging sequences because accurate characterization of lesions is critical for effective patient treatment. Gadolinium, however, is a cause for concern among clinicians due to the element's association with nephrogenic systemic fibrosis and evidence of gadolinium deposition in various tissues and bone long after GBCA-enhanced scans.
One potential solution is deep learning, which was used in a previous study to successfully elevate the detection of active MS lesions. For the current study, the researchers took deep learning one step further by using the artificial intelligence approach "to identify enhancing lesions on unenhanced images," they noted.
"The ability to identify enhancing lesions without GBCA administration by using [deep learning] can potentially minimize the need for GBCA administration, improve patient safety, and reduce costs associated with clinical care," they wrote.
The authors prospectively collected multiparametric MR images from 1,008 relapsing-remitting MS patients (mean age, 37.7 ± 9.7 years; range, 18 to 60 years) who underwent a total of 1,970 1.5- or 3.0-tesla scans as part of a randomized phase III clinical trial at one of 68 imaging centers. The MRI protocol included 2D FLAIR and T1-weighted sequences.
"In this feasibility study, we included images from the baseline examination and the follow-up examination at six months," Narayana and colleagues explained. "MRI scans in this cohort were also analyzed to determine the regional atrophy, effect of inpainting, effect of intrinsic and extrinsic factors on cortical thickness, and tissue segmentation."
The researchers trained a convolutional neural network to classify enhanced lesions on unenhanced 2D MRI slices for possible contrast enhancement in each slice. They then employed a connected network to combine the slice scores to accurately predict results.
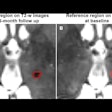
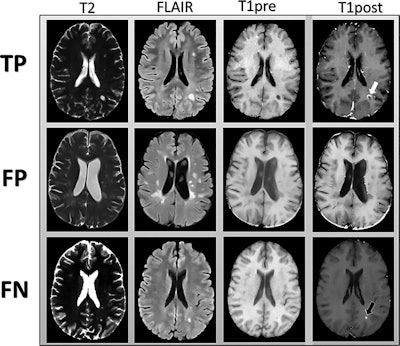 MR images are examples of T2-weighted, fluid-attenuated inversion-recovery (FLAIR), and precontrast T1-weighted images input into the deep-learning model. Postcontrast T1-weighted images (T1post) demonstrate areas of true-positive (white arrow) and false-negative (black arrow) enhancement. FN = false-negative classification of enhancement, FP = false-positive classification of enhancement, TP = true-positive classification of enhancement. Images courtesy of Radiology.
MR images are examples of T2-weighted, fluid-attenuated inversion-recovery (FLAIR), and precontrast T1-weighted images input into the deep-learning model. Postcontrast T1-weighted images (T1post) demonstrate areas of true-positive (white arrow) and false-negative (black arrow) enhancement. FN = false-negative classification of enhancement, FP = false-positive classification of enhancement, TP = true-positive classification of enhancement. Images courtesy of Radiology.Their deep-learning approach achieved a sensitivity of 78% (± 4%) and specificity of 73% (± 2.7) averaged across the five test sets for predicting contrast enhancement in each MRI slice. In addition, sensitivity was 72% (± 9%) and specificity was 70% (± 6%) in determining the presence of any enhancing lesions in an MS patient.
"Even though we used data acquired at different centers with different MRI system platforms, the MRI parameters were well controlled in this clinical trial," the researchers added. "For generalizability of the model, it is essential that further testing be conducted on a heterogeneous dataset."


.fFmgij6Hin.png?auto=compress%2Cformat&fit=crop&h=100&q=70&w=100)




.fFmgij6Hin.png?auto=compress%2Cformat&fit=crop&h=167&q=70&w=250)