Deep learning could improve real-time lung ultrasound interpretation, according to a study published January 29 in Ultrasonics.
Researchers led by Lewis Howell, PhD, from the University of Leeds in the U.K. found that a deep learning model trained on lung ultrasound allowed for segmentation and characterization of artifacts on images when tested on a phantom model.
“Machine learning and deep learning present an exciting opportunity to assist in the interpretation of lung ultrasound and other pathologies imaged using ultrasound,” Howell and colleagues wrote.
Lung ultrasound in recent years has been highlighted in research as a safe, cost-effective imaging modality for evaluating lung health, and the COVID-19 pandemic saw lung ultrasound utilized more as a noninvasive imaging method.
Still, the researchers noted that this method has its share of challenges, especially user variability.
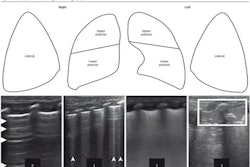
Howell and co-authors described a deep learning method for multi-class segmentation of objects such as ribs and pleural lines, as well as artifacts including A-lines, B-lines, and B-line confluences in ultrasound images of a lung training phantom.
The team developed a version of the U-Net architecture for image segmentation to provide a balance between model speed and accuracy. It also used an ultrasound-specific augmentation pipeline during the training phase to improve the model’s ability to generalize unseen data, including geometric transformations and ultrasound-specific augmentations.
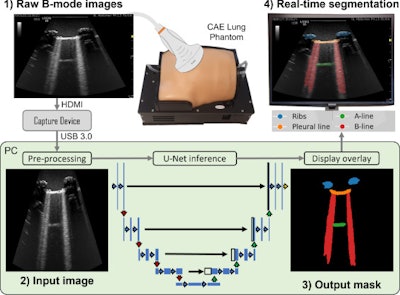 Workflow diagram showing real-time lung ultrasound segmentation with U-Net. Images available for republishing under Creative Commons license (CC BY 4.0 DEED,
Workflow diagram showing real-time lung ultrasound segmentation with U-Net. Images available for republishing under Creative Commons license (CC BY 4.0 DEED,
Attribution 4.0 International) and courtesy of Ultrasonics.
Finally, the researchers used the trained network to segment a live image feed from a cart-based point-of-care ultrasound (POCUS) system (Venue Fit R3, GE HealthCare, or GEHC). They also imaged the training phantom using a convex curved-array transducer (C1-5-RS, also GEHC) and streamed frames.
The team trained the model on a single graphics processing unit, which took about 12 minutes with 450 ultrasound images.
The model achieved an accuracy of 95.7%, as well as moderate-to-high Dice similarity coefficient scores. The investigators added that applying the model in real-time, at up to 33.4 frames per second in inference, improved the visualization of lung ultrasound images.
| Performance of deep-learning model on training phantom | |
|---|---|
| Metric | Dice score |
| Background | 0.98 |
| Ribs | 0.8 |
| Pleural line | 0.81 |
| A-line | 0.63 |
| B-line | 0.72 |
| B-line confluence | 0.73 |
Additionally, the team assessed the pixel-wise agreement between manually labeled and model-predicted segmentation masks. Using a normalized confusion matrix for precision, it found that of the manually labeled images, the model correctly predicted 86.8% of pixels labeled as ribs, 85.4% of pleural line, and 72.2% of B-line confluence. However, only 57.7% of A-line and 57.9% of B-line pixels were correctly predicted by the model.
Finally, the researchers used transfer learning for their model, which leverages knowledge gained through training on one dataset to inform training on a related dataset. They found that the per-class Dice similarity coefficients for simple pleural effusion, lung consolidation, and pleural line were 0.48, 0.32, and 0.25, respectively.
The study authors suggested that their model could assist with lung ultrasound training and in addressing skill gaps. They proposed a semiquantitative measure called the B-line Artifact Score, which is related to the percentage of an intercostal space occupied by B-lines, which could be tied to the severity of lung conditions.
“Future work should consider the translation of these methods to clinical data, considering transfer learning as a viable method to build models which can assist in the interpretation of lung ultrasound and reduce inter-operator variability associated with this subjective imaging technique,” they wrote.
The full study can be found here.