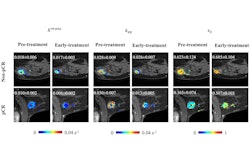
A deep-learning system used with dynamic contrast-enhanced MRI (DCE-MRI) could serve as a predictive tool in measuring pathological complete response in breast cancer patients, suggest findings shared at the recent RSNA meeting.
In his presentation, Yi Dai, MD, PhD, from Peking University Shenzhen Hospital in China discussed the research results, showing that the deep-learning system achieved high marks in prospective testing and other tests.
“The … system presents a fully automated, non-invasive, and reliable tool for early pathologic complete response prediction in breast cancer patients undergoing neoadjuvant chemotherapy,” Dai told session attendees.
 Yi Dai, MD, PhD, from Peking University Shenzhen Hospital in China discussed his team's findings at RSNA 2024, showing that a fully automated deep-learning system based on dynamic contrast-enhanced MRI can predict pathological complete response in breast cancer patients undergoing neoadjuvant chemotherapy.
Yi Dai, MD, PhD, from Peking University Shenzhen Hospital in China discussed his team's findings at RSNA 2024, showing that a fully automated deep-learning system based on dynamic contrast-enhanced MRI can predict pathological complete response in breast cancer patients undergoing neoadjuvant chemotherapy.
Neoadjuvant chemotherapy is the standard treatment method for women with locally advanced breast cancer. Women who achieve pathological complete response have more favorable outcomes. This makes accurate prediction of complete response important for optimal treatment.
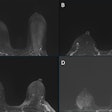
Dai said that DCE-MRI has advantages over mammography and ultrasound in this area by providing more comprehensive tumor information. But the modality is limited by its complexity and heterogeneity of response, as well as high variability in image interpretation by radiologists.
Dai and colleagues developed and sought to validate a fully automated deep-learning system by using the pre-enhanced and peak enhanced phases images of DCE-MRI. They intended for the system to predict pathological complete response to neoadjuvant chemotherapy in breast cancer. The team also explored the biological basis underlying prediction by the deep-learning system.
The study included 4,041 women who underwent DCE-MRI before chemotherapy from 10 medical centers and from data in The Cancer Genome Atlas Program. The women were divided into training, internal testing, external testing, and prospective testing sets.
The researchers also conducted RNA sequencing and genetic analysis on 100 women to explore the biological basis of the deep-learning system.
The system achieved high marks on all test groups. This included the following: training set, area under the curve (AUC) = 0.853; internal testing set, AUC = 0.850; external testing set, AUC range from 0.807 to 0.834; and prospective testing set, AUC = 0.833.
Also, genetic analysis showed that high scores given by the deep-learning system were tied to up-regulation of immune-mediated genes and pathways.
Dai said that with these results in mind, the fully automated deep-learning model can aid clinicians in forming and implementing personalized treatment plans for patients.
“Moreover, the underlying biological basis of deep-learning for pathological complete response prediction to neoadjuvant chemotherapy might be related to the up-regulation of immune-mediated genes and pathways,” Dai said.