3D anatomical modeling using advanced imaging software can be cost-effective in planning pediatric surgeries, according to research published October 28 in the Journal of Pediatric Surgery.
A team led by Mark Ryan, MD, from the University of Texas Southwestern Medical Center in Dallas found that its modeling approach can successfully visualize pediatric anatomical structures. From there, these 3D structures can be retained for 3D printing, virtual reality usage, or as an anatomic reference.
“Visualization and preoperative planning can be assisted by advanced imaging software at minimal to no cost, thereby facilitating enhanced understanding of these conditions in resource-limited environments,” the Ryan team wrote.
Pediatric surgeons often must strategize treatment for complex congenital abnormalities and other conditions that require a thorough understanding of the conditions' anatomy. The researchers noted that there are methods available to transfer cross-sectional imaging data into 3D models for education and preoperative planning purposes, but that surgical centers, especially those with limited resources, may be discouraged from using them due to the associated costs and lack of familiarity.
Ryan and co-authors sought to describe a low-cost, reproducible method for generating 3D images to visualize pediatric patient anatomy.
The team obtained deidentified DICOM files from a hospital PACS system used to prepare for pediatric surgical procedures. From there, it used open-source visualization software to analyze variations in anatomical structures, which used volume rendering and segmentation techniques. Finally, the researchers used editing tools and AI-assisted software extensions to further refine generated images.
The researchers described the model’s use in three cases.
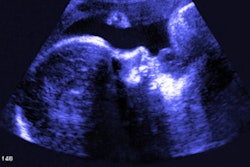
One was for confirmation of a choledochal cyst in a 4-year-old girl. An MR cholangiopancreatography image was uploaded into 3D Slicer, from which the team identified a type I choledochal cyst. With image rotation, it could view aspects of the cyst that had not been mentioned in initial imaging scans.
“The contrast permitted easy segmentation of the bile ducts and gallbladder using the segmentation editor tool,” the researchers wrote. “The accessory duct could be easily visualized, and the models were then inserted into the volume rendering for viewing in virtual reality.”
The patient subsequently underwent laparoscopic resection of the cyst and hepaticoduodenostomy.
In a second case, the model was used to view volume renderings of a congenital pulmonary airway malformation in a male infant born at 36 weeks' gestation. From here, the researchers viewed the vascular supply through a 3D transparent lung window and created a model from segmentation of the lung lesion and airways. An AI segmentation extension to Slicer 3D was then used to automatically model the remaining lobes of the lung, and the editing tool was used to adjust their opacity.
The models were then added to the volume rendering and on rotation of the view the aortic feeding vessels could be well visualized. The patient was electively taken to the operating room for thoracoscopic left lower lobectomy, the team reported.
Finally, in a third case, Slicer 3D was used for treatment planning of a four-year-old female patient with a renal mass. An AI extension to the model identified and further modeled the anatomic structures in a CT scan in about 45 seconds.
From there, the team performed manual segmentation and with rotation identified “almost complete” obliteration of the right hepatic vein and determined the tumor’s location. The patient underwent neoadjuvant chemotherapy.
The model can operate on any personal computer released in the last five years, the authors wrote, though they recommended that dedicated graphics hardware be used for volume rendering.
They also highlighted that using AI tools for image recognition and volume reconstruction could reduce barriers to entry and improve access to complex reconstructions of pediatric surgical anatomy.
The full report can be found here.