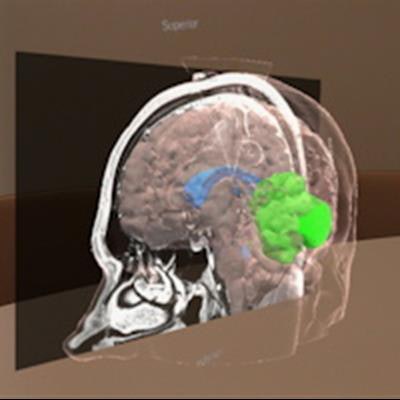
An automated MR image segmentation algorithm can generate 3D models of brain tumors that neurosurgeons can use for augmented reality (AR)-based viewing and surgical planning, according to research published in the Journal of Neurosurgery.
Researchers led by Dr. Tim Fick of the Princess Máxima Center for Pediatric Oncology in Utrecht, the Netherlands, developed and validated a cloud-based segmentation algorithm that automatically generates 3D models from contrast-enhanced T1-weighted MR sequences. These models are then automatically optimized and prepared for neurosurgeons to view in 3D on a computer or AR headset. In testing on 50 patients with a brain tumor, their approach was highly accurate and much faster than manual segmentation.
"The automatic cloud-based segmentation algorithm is reliable, accurate, and fast enough to aid neurosurgeons in everyday clinical practice by providing 3D augmented reality visualization of contrast-enhancing intracranial lesions measuring at least 5 cm," the authors wrote.
A number of studies have shown the benefits of 3D models and AR in visualizing complex pathology and anatomy, as well as for surgical planning and preparation. However, this currently requires multiple manual steps, including exporting DICOM images from the PACS, creating 3D models with manual or semiautomated segmentation, optimizing the models for visualization, simplifying the models to account for limited processing power, and then saving them into different file formats. This process is cumbersome and therefore can't be integrated into daily clinical practice, according to the researchers.
"To make this technique more accessible to surgeons, we built a fully automatic workflow that uses DICOM images to create 3D models suitable for AR-[head-mounted displays] and is integrated with an automatic segmentation algorithm that segments skin, brain, ventricles, and tumor," the authors wrote.
The entire process is conducted via a cloud-based application called AugSafe from AR software developer Augmedit. The resulting 3D models can be viewed either on a web-based interface on a computer, as a holographic scene on an AR head-mounted display such as a Microsoft HoloLens, or other devices using an automatically generated QR code, according to the researchers.
![3D models generated with automatic segmentation and shown through an embedded 3D viewer in a web-based interface (upper) and an AR-[head-mounted display] (lower). Images and caption courtesy of the Journal of Neurosurgery.](https://img.auntminnie.com/files/base/smg/all/image/2021/08/am.2021_08_12_22_05_4265_2021_08_19_AR_Surgery.png?auto=format%2Ccompress&fit=max&q=70&w=400) 3D models generated with automatic segmentation and shown through an embedded 3D viewer in a web-based interface (upper) and an AR-[head-mounted display] (lower). Images and caption courtesy of the Journal of Neurosurgery.
3D models generated with automatic segmentation and shown through an embedded 3D viewer in a web-based interface (upper) and an AR-[head-mounted display] (lower). Images and caption courtesy of the Journal of Neurosurgery.In addition, the 3D models can be viewed within the original DICOM images, as well as manipulated and viewed from different angles.
The researchers compared the performance of their technique with manual image segmentations -- performed on the open-source 3D Slicer software -- in 50 patients with a brain tumor volume of 5 cm3 or more that had been treated between 2009 and 2020 at the University Medical Center Utrecht.
| Performance of AugSafe algorithm for automated image segmentation | |||
| Mean dice similarity coefficient | 0.87 ± 0.07 | ||
| Average symmetric surface distance | 1.31 ± 0.63 mm | ||
| 95th percentile of Hausdorff distance | 4.80 ± 3.18 mm | ||
| Segmentation time | 753 ± 128 seconds | ||
In comparison, manual segmentation took a mean of 6,212 ± 2,039 seconds and only included the tumor.
The authors acknowledged the limitations of the algorithm with regards to a minimum tumor volume and the pathologies it can be used for.
"The next steps involve incorporation of other sequences and improving accuracy with 3D fine-tuning in order to expand the scope of augmented reality workflow," they wrote.