How many radiologist-annotated images are needed to train a deep-learning algorithm for radiology applications? Perhaps not as many as you might think. Using both annotated and unannotated images, researchers from Google have developed an algorithm that can simultaneously identify and localize disease found on chest x-rays.
Jia Li, head of R&D, cloud AI, and machine learning at Google, highlighted the algorithm's potential to help radiologists interpret chest x-rays during a presentation on March 27 at MIT Technology Review's EmTech Digital conference in San Francisco.
"It outperforms the state-of-the-art machine-learning technology [for] disease prediction, and more importantly, it generates insights about the decision that has been made [by the model] to assist better interpretability [by the radiologist] of the result," Li said.
Chicken and egg problem
Chest x-rays remain a significant challenge in radiology, requiring extensive expertise by radiologists to accurately interpret these images, Li said.
"If we could build AI power into radiology image analysis, we can help to assist [radiologists] to make even better sophisticated judgments, and make the entire process much more efficient," she said.
There's a chicken and an egg problem, though, Li said.
"On one hand, radiologists are spending a lot of time analyzing the radiology images, and we want to build AI-powered tools to assist their diagnosis," she said. "But in order to do so, we need a large amount of labeled data. That goes back to the exact problem that we want to solve and puts [the burden back on] our radiologists to label a large amount of data."
Identifying, localizing disease
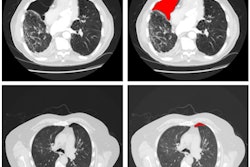
In an attempt to solve this problem, the researchers developed a deep-learning model -- trained using a limited number of annotated images -- that can simultaneously identify disease and localize it on the image for the radiologist to see.
"We propose a new approach by combining the overall disease types and the local pixel-wise indication of disease without additional detailed labeling effort," Li said. "The resulting solution generates overall prediction of the disease type as well as [the location of] the abnormal area of the disease."
In a paper published on arXiv.org, the researchers -- led by first author Zhe Li (now at Syracuse University) -- shared how they began their effort by utilizing the U.S. National Institutes of Health (NIH) ChestX-ray8 database, an open-source dataset of more than 110,000 chest x-ray studies that have been associated with up to 14 disease types. This dataset also includes 880 images with 984 "bounding boxes," or annotated regions of an image that show a disease, that were provided by board-certified radiologists. These annotated cases encompassed eight disease types.
Annotated, unannotated cases
To train the model, the researchers used the 880 images with annotations and the remaining 111,240 images without annotations, which were labeled as having a particular disease but did not include specific localization of the disease on the image. In their approach, a convolutional neural network (CNN) is applied to an input image to classify the entire image information and to encode both the class (i.e., type of disease) and localization information. The image is then sliced into a "patch" grid to capture the local information of the disease, according to the researchers.
"For an image with bounding box annotation, the learning task becomes a fully supervised problem since the disease label for each patch can be determined by the overlap between the patch and the bounding box," they wrote. "For an image with only a disease label, the task is formulated as a multiple instance learning (MIL) problem -- at least one patch in the image belongs to that disease."
As a result, the processes of disease identification and localization are combined in the same underlying prediction model, according to the group.
After training was completed, the researchers compared the performance of their model on the NIH dataset with Res-Net-50, a deep-learning algorithm developed by an NIH team. After performing receiver operating characteristic analysis, they found that their deep-learning method yielded a higher area under the curve (AUC) than the NIH algorithm for detecting 14 conditions on the chest x-rays.
| Comparison of algorithms for detecting findings on chest x-ray | ||
| Conditions | AUC of NIH Res-Net-50 | AUC of Google model |
| Atelectasis | 0.72 | 0.80 ± 0.00 |
| Cardiomegaly | 0.81 | 0.87 ± 0.01 |
| Consolidation | 0.71 | 0.80 ± 0.01 |
| Edema | 0.83 | 0.88 ± 0.01 |
| Effusion | 0.78 | 0.87 ± 0.00 |
| Emphysema | 0.81 | 0.91 ± 0.01 |
| Fibrosis | 0.77 | 0.78 ± 0.02 |
| Hernia | 0.77 | 0.77 ± 0.03 |
| Infiltration | 0.61 | 0.70 ± 0.01 |
| Mass | 0.71 | 0.83 ± 0.01 |
| Nodule | 0.67 | 0.75 ±0.01 |
| Pleural thickening | 0.71 | 0.79 ± 0.01 |
| Pneumonia | 0.63 | 0.67 ± 0.01 |
| Pneumothorax | 0.81 | 0.87 ± 0.01 |
"Our quantitative results show that the proposed model achieves significant accuracy improvement over the published state-of-the-art on both disease identification and localization, despite the limited number of bounding box annotations of a very small subset of the data," the authors wrote. "In addition, our qualitative results reveal a strong correspondence between the radiologist's annotations and detected disease regions, which might produce further interpretation and insights of the diseases."
Li noted that Google is taking baby steps in developing AI for healthcare applications.
"For example, partners such as Zebra Medical are using Google Cloud to analyze new scans and inform insights to hospitals to make better decisions at scale," she said. "There is still much for us to do in this space. Hopefully, in the future, specialists will spend less time on repetitive and error-prone tasks with the assist of AI technology."