Thursday, November 29 | 11:10 a.m.-11:20 a.m. | SSQ11-05 | Room S103AB
Imaging annotations stored in a proprietary fashion can be converted into a standardized format, enabling interoperability and automated lesion matching across sequential imaging studies, according to this presentation.Although the ability to access and share radiologic imaging annotations is important in a number of clinical and research contexts, the tendency of commercial PACS vendors to encode annotations in their own proprietary formats -- often with DICOM presentation state objects -- prevents these data from being readily accessed, noted presenter Dr. Nathaniel Swinburne from Memorial Sloan Kettering Cancer Center in New York City.
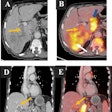
To help with this situation, the researchers created software to extract line annotations encoded in a proprietary format on version 3.x of the Centricity PACS software (GE Healthcare) and then convert them to the standard Annotation and Image Markup (AIM) format. They then demonstrated the utility of this conversion by performing automated matching of lesion measurements across time points for cancer lesion tracking.
To validate their approach, the researchers exported anonymized DICOM images and DICOM presentation state objects containing lesion annotations from the PACS and processed them with their software. Next, they imported the DICOM files along with the matched AIM annotation files into the freely available electronic Physician Annotation Device (ePAD) AIM-compliant workstation software.
The matched lesion measurement annotations were correctly linked by ePAD across sequential imaging studies, enabling automatic quantitative evaluation of tumor response via functionality already existing in ePAD, Swinburne said.
"Our work demonstrates that proprietary image annotations in a vendor system can be automatically converted to a standardized metadata format such as AIM, enabling interoperability and potentially facilitating large-scale analysis of image annotations and the generation of high-quality labels for deep-learning initiatives," Swinburne told AuntMinnie.com. "In the future, this effort could be extended for use with other vendors' PACS."