The digitization of radiology has made the concept of efficient, cost-effective data mining possible, while the development of informatics tools is making such activity realistic. Two articles in the August issue of Radiology describe tools to extract radiation dose data.
A team of radiologists and radiology informatics subspecialists at Brigham and Women's Hospital have developed two open-source informatics toolkits. One extracts anatomy-specific CT radiation exposure metrics, while the other captures data on radiopharmaceuticals and administered activities from nuclear medicine exam reports. With each, the data can be used for radiation monitoring of patients and in quality assurance programs (Radiology, August 2012, Vol. 264:2, pp. 397-405 and pp. 406-413).
Nuclear medicine informatics tool
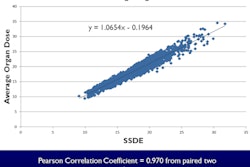
Nuclear medicine exams account for approximately 26% of the radiation dose from medical imaging performed in the U.S. Radiation dose to organs may be calculated with knowledge of the radiopharmaceutical used and how it was administered. Information is contained in the text of radiology reports. Unlike CT imaging, radiation dose to individual organs is not a factor of the size of the patient.
Lead author Dr. Ichiro Ikuta, a postdoctoral fellow at the Center for Evidence-Based Imaging, and colleagues designed an open-source toolkit with the ability to extract radiation exposure data from radiology reports contained in Brigham and Women's RIS and import the information into database and spreadsheet software. The Perl Automation for Radiopharmaceutical Selection and Extraction (PARSE) converts all units of radioactivity into a standardized format, with each identified unit of radioactivity used as an anchor point to search for numeric expressions of administered activity within a set text boundary. Broader searches from each unit of radioactivity are performed to match a user-defined list of radiopharmaceutical products.
If a report lacks a unit of radioactivity or does not have content in its radiation exposure data fields, it is identified as "null" and "uncertain," respectively. These reports may be used in a quality control program to determine the frequency of incomplete data and the source.
To validate the toolkit, the research team identified all nuclear medicine reports prepared between September 1985 and March 2011. In less than 11 minutes, 204,561 reports were identified, from which 5,000 reports were randomly selected and used to train the software.
A portion of the 5,000 reports were used for manual validation of PARSE. The remainder (2,359 reports) were analyzed by PARSE to identify how many and which contained information about a single radiopharmaceutical with a single or multiple administration, and multiple radiopharmaceuticals with multiple administrations. Ninety percent (2,131 reports) were complete and analyzed by the researchers for recall -- specifically, the percentage of complete reports from which all data fields were correctly extracted (97.6%) -- and for the precision of data extraction -- specifically, the percentage of reports with extracted data that were correct (98.7%).
Lack of a named radiopharmaceutical was the most common recall error (63%), followed by typographical errors and unique formatting variations in the report. The researchers identified two causes for precision errors: double counting of units of radioactivity caused by an extra field containing this information, and unique report formats. They have expanded the library of radiopharmaceutical agents and identified methods of flagging reports that indicate an "extra dose" entry or cumulative dose calculations.
For quality assurance programs, Ikuta and colleagues suggested that predefined acceptable ranges of data could be identified, to which outlier values in reports could be compared. They also recommended that the data repository could be used to monitor exam protocol changes over time, something they demonstrated in their evaluation. With respect to patients, because radiopharmaceutical administration can be mapped to organ doses, longitudinal patient-specific timelines and cumulative organ dose heat maps can be developed.
CT exam radiation exposure extraction tool
The generalized radiation observation kit (GROK) is an open-source toolkit that extracts anatomy-specific CT radiation exposure metrics (CT dose index volume and dose-length product) from existing digital image archives. To do this, it uses optical character recognition of CT dose report screen captures and DICOM image attributes containing information about the CT scanner, CT protocol, type of examination, and patient demographics. The toolkit performs automated anatomic assignment of exposure metrics, enabling comparison among radiology facilities that use a variety of different protocol names for the same category of exam being performed.
Lead author Dr. Aaron Sodickson, PhD, associate professor of radiology at Harvard Medical School, and colleagues at Brigham's Center for Evidence-Based Imaging validated the extraction and anatomy assignment algorithms by sampling all CT studies performed during a specific week each quarter of calendar years 2000 through 2010. They obtained 54,549 CT encounters of patients who were imaged by scanners manufactured by GE Healthcare, Philips Healthcare, Siemens Healthcare, and Toshiba America Medical Systems.
The process of extracting data took approximately 110 hours, at a rate of 50 CT studies per hour. Fifty-five percent of these had at least one detected dose screen. This number increased dramatically, from only 10% of exams performed in the first quarter of 2002 to 95% in the fourth quarter of 2010, reflecting the purchase of new CT scanners and implementation of software upgrades.
Overall, the dose screen retrieval rate was 99% and the anatomic assignment precision rate was 91%. All errors represented failure to assign the correct anatomic region to the dose event. Naming errors comprised 83% of the error rate; these were associated with inadequate series and protocol descriptions. Matching errors (13%) and optical character recognition errors (4%) were the other types of errors identified.
The toolkit is strictly an extraction tool and does not contain software to analyze data. The authors pointed out that many radiology departments might not have the staff resources to provide this. They also noted that while the extracted data are x-ray tube metrics, they are not correlated with patient size metrics, information that is not yet routinely available in existing archives. Another concern they identified is the potential need for repeated toolkit customization efforts when manufacturers change dose screen format and contents.
"Automatic extraction of anatomy-specific exposure data does not in itself solve exposure monitoring problems, and considerable effort is required to implement supporting policies and transform associated processes to realize the benefits enabled by this new technology," they cautioned. However, its use could greatly benefit both radiation exposure quality assurance initiatives and imaging technique optimization programs for CT exams.
Enabling practice-specific auditing
In an accompanying editorial, Cynthia McCollough, PhD, professor of biomedical engineering and medical physics at the Mayo Clinic in Rochester, MN, praised the development of these toolkits. Such tools can make practice-specific auditing of radiation dose levels possible, which is needed to adequately monitor the performance of a department and to benchmark against others on a facility, regional, national, or international level.
"The availability of data describing the amount of radiation used for different examinations or procedures, stratified by patient size and clinical indication, is foundational for quality improvement initiatives in the field of radiation dose utilization," she wrote.
McCollough commended the American College of Radiology (ACR) for creating its Dose Index Registry and providing a source of benchmark information as well as baseline data to measure practice improvements. But she noted that having a large sample of dose index values is not enough.
Comparing CT dose index values of an obese patient with those of a thin adult may unfairly reflect dose variation in a department. Noting that each dose index value must be associated with not only a specific anatomic region but also a specific patient size and diagnostic task, McCollough reiterated that an automated measure of patient size needs to be standardized and stored in a DICOM header field.
In addition to establishing stratification for anatomic regions, McCollough also emphasized the importance of developing a system to specify the diagnostic task of an exam. As an example, she cited dose differences between a lung cancer screening exam and imaging the thorax to identify pulmonary emboli.
"One of the exciting capabilities demonstrated [by the Brigham team] is the ability to examine dose metrics across time; between facilities, manufacturers, and scanner models; between specific protocols; and even for a specific patient," she wrote. "The richness of the data provided and the potential uses for quality improvement initiatives are compelling and should motivate professional societies, standard organizations, regulators, and manufacturers to adopt standardized measures of patient size, anatomic scan region, and diagnostic task as quickly as possible."