Developers of an advanced visualization algorithm known as Virtual Finger (VF) are hoping to revolutionize the tedious and slow process of exploring 3D image content. While the algorithm is currently being employed for nonmedical applications, its developers believe it could be used clinically in the near future.
3D imaging has seen great progress in recent years, but finding and analyzing specific places in a 3D volume is still an arduous task -- one that forces advanced imaging research into the slow lane and keeps many clinical applications unfeasible, according to researchers at the Allen Institute for Brain Science.
Virtual Finger, in development by Hanchuan Peng, PhD, and colleagues at the institute for several years, promises to open new doors to the possible with a virtually instantaneous software technique that zooms in to any spot the user might want to click on, enabling fast exploration of complex 3D content.
 Hanchuan Peng, PhD, from the Allen Institute for Brain Science.
Hanchuan Peng, PhD, from the Allen Institute for Brain Science.To date, the developers haven't focused much energy on medical imaging applications, but the software is a natural fit for 3D image interpretation in radiology or intraoperative use in image-guided surgery, Peng told AuntMinnie.com.
"Medical imaging is actually pretty straightforward as long as 3D data have been collected," he said. "In MRI of the brain, there are a lot of structures, for example, the brain vasculature. How can you actually reconstruct blood vessels in real-time during a surgical procedure? If you want to insert something in the brain or do some sort of gamma ray-based surgery using a Gamma Knife, you will have to first find out where you want to target."
Virtual Finger can help with that decision. One need only display the entire volume of data in front of the physician and the software can instantly generate a 3D target. (A YouTube video clip offers an overview of the technique.)
"You save time, and the saved time could be critical for many purposes," Peng said. Maybe the dataset is large and the surgical planning involves many sites. Going through all of them slice by slice takes an excessive amount of time.
"Now it's become very simple: You just click on your screen and you're done," he said.
Virtual Finger is the only technique that allows researchers to start with a 2D image and then recover the missing dimension with other software, whether the purpose is for medical imaging or the video gaming industry, Peng said. And although the results are fast, the math behind it is complex.
On a 2D monitor, you can't really directly operate on 3D content. You need an alternative approach, according to Peng. Conventional 3D software works by dividing 3D images into thin sections that can be viewed on a flat screen. To access that content, the reader must go through the volume and predefine individual sections, but given the sheer volume of today's imaging datasets, it's a painfully slow process.
"That's why all of the applications are labor-intensive," he said. "The new technique is basically what you see in 2D with a simple operation of your mouse or finger on a touchscreen -- you just move your finger to move around the object, and the software is able to translate such prior 2D information back to the 3D space and give you the missing dimension. That's why we call the technique the Virtual Finger, and that kind of ability was not available before."
Turning selected pieces of 2D image content back into 3D has defied easy solutions, Peng and colleagues wrote in an article that appeared this summer in Nature Communications (July 11, 2014). Previous algorithms have accomplished the task of pinpointing 2D spaces and converting them to 3D, later even creating curves from data points, but they all operated slowly and were prone to crashing.
Virtual Finger instead represents a family of open-source computing methods that "generate 3D points, curves, and region-of-interest (ROI) objects in a robust and efficient way," the authors wrote in the July report. As long as the objects are visible on a 2D display device, a single mouse click or command from another input device, such as a touchscreen, allows Virtual Finger to reproduce 3D locations in the image volume.
The technology allows researchers to virtually reach into the digital space of objects as small as a single cell. The Nature Communications article discussed several applications of the technology in nonmedical imaging, along with image-related procedures that encompass "data acquisition, visualization, management, annotation, analysis, and use of the image data for real-time experiments such as microsurgery, which can be boosted by 3D-VF," the authors wrote.
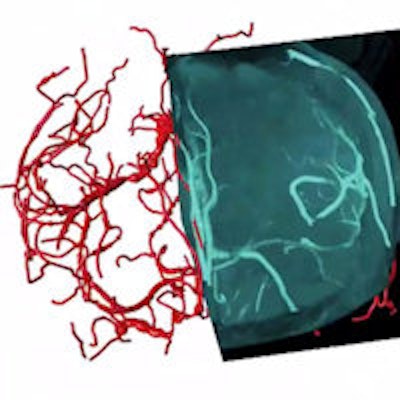
3D vasculature of the brain extracted from an MRI volume. Virtual Finger was used to both define the initial tracing locations and edit the result in a fraction of the time required to perform the operations manually. Video courtesy of Hanchuan Peng, PhD.
The paper highlights three case studies that involve 3D imaging, profiling, and free-form in vivo microsurgery in fruit flies and roundworms. A second experiment performed instant 3D visualization and annotation of "terabytes of microscopic image volume data of whole mouse brains." A third experiment demonstrated efficient reconstruction of complex neuron morphology from images of dragonfly and other systems that present high levels of noise, "enabling the generation of a projectome of a Drosophila [fruit fly] brain," the authors wrote.
For looking at single cells of insects, all you need is a digital camera, Peng said. You acquire a larger view at low resolution, use it to find the region you want, and zoom in for a higher resolution image. Then, when you point to a specific target, the Virtual Finger can transform it into a 3D picture.
Eyes have it
The process used by the VF algorithm is computationally very efficient and occurs in milliseconds, because finding the 3D location of an object is based on projected rays going in slightly different angles. Conceptually, it is akin to a pair of human eyes being far enough apart to see slightly different views, Peng explained.
"The key idea is that you can generate many rays, and you can march through the rays with the algorithm -- it's close to what humans see," he said.
Human vision is essentially the 3D perception of 2D projections.
"The whole reason why you can perceive some sort of 3D image in your brain is because there is some sort of external intersection strategy using both your eyes," he said. "You have a left eye and a right eye, and your brain is going to create a 3D image."
Indeed, the algorithm, known as CDA2, employs an adapted fast-marching algorithm to search for a shortest geodesic path from the last knot on the curve, each of which has a 3D location, to the current ray. It then iterates this process to complete an entire curve.
The algorithm provides additional flexibility by enabling methods to refine the 3D curves using extra mouse operations.
"CDA2 is robust in generating consistent and precise 3D curves from different viewing angles, zooms, and other perspective parameters of an image," Peng and colleagues wrote.
The algorithm also uses a machine learning approach to profile the data and create a kind of prior, in a process similar to accessing an imaging atlas.
"You get the most probable location of an object," he said. "You do not need to use an intersection strategy anymore, and that saves you one eye. That's the key to efficiency."
Peng said he actually did most of the development work on the algorithm several years ago at the Howard Hughes Medical Institute; he then continued the work over the past two years at the Allen Institute. Commercialization efforts are underway, but details of any collaboration can't be disclosed yet, he said.
The technique could make data collection and analysis 10 to 100 times faster than any current method, and render innumerable kinds of imaging research feasible, Peng said.
"This is kind of like unexplored territory, so it was likely unknown that it could ever be made so fast," he said.