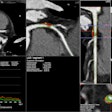
Researchers at University of California, San Francisco (UCSF) and Duke University Medical Center in North Carolina have created an automatic deep learning-based neonatal brain MRI extraction data algorithm.
The deep learning (DL) algorithm, called ANUBEX, performs automatic segmentation, also called skull-stripping, of neonatal brains from MR images. Model training was accomplished using an iterative, human-in-the-loop AI approach. The article was published February 26 in Nature Scientific Reports.
What drives this project is that brain extraction is well established with adult brain models but not so for the neonatal brain, explained Joshua Chen, MD, from the UCSF Department of Radiology, and collaborators that included Evan Calabrese, MD, PhD, from the Duke Center for Artificial Intelligence in Radiology. The deep-learning approach addresses a need for improved MRI brain extraction for newborn brains along with generalizability of AI algorithms.
Designed as an automated neonatal nnU-Net brain MRI extractor, ANUBEX was developed using T1-weighted, T2-weighted, and diffusion-weighted imaging (DWI) from neonatal MRI studies. Scan parameters varied based on the imaging site and scanner platform, according to the authors.
The study population consisted of newborns with moderate to severe encephalopathy at birth who were part of the High-dose Erythropoietin for Asphyxia and Encephalopathy (HEAL) study. With informed consent previously obtained, participants were prospectively enrolled from 17 different institutions across the U.S. Exclusion criteria included missing, incomplete, or severely artifact-degraded T1-weighted MR imaging data.
"First, baseline automated brain masks were generated from T1-weighted images using a widely used tool for adult MRI brain extraction," the authors wrote. "Next, all brain masks were manually reviewed by a single medical trainee (author JC) using ITK-SNAP and categorized. ... Final model training using all the manually reviewed/corrected data (N = 433) was performed using a fivefold cross-validation approach with a standard random 80%/20% train/validation split for each fold. Model training was accomplished using a desktop computer equipped with two Nvidia RTX A600 40 GB graphics processing units running in parallel (one training fold per GPU)." Final model training lasted approximately 36 hours.
For external validation of ANUBEX, the researchers used an external sample set from the developing Human Connectome Project, a database that houses public research data and supports numerous studies that focus on the connections within the human brain.
Model performance was compared with five different publicly available automated brain extraction methods: BET, BSE, CABINET, iBEATv2, and ROBEX. The researchers also evaluated the model in various scenarios, such as preterm brain MRIs and motion-degraded brain MRIs.
"ANUBEX performs similarly when trained on sequence-agnostic or motion-degraded MRI, but slightly worse on preterm brains. In conclusion, we created an automatic deep learning-based neonatal brain extraction algorithm that demonstrates accurate performance with both high- and low-resolution MRIs with fast computation time," the authors stated.
The authors cited their iterative semiautomated approach to ground truth brain mask generation as helpful for the efficiency and consistency of their deep-learning algorithm. In addition, the combination of using a multi-institutional training dataset from the HEAL trial and including variation in MRI manufacturer, model, software, and imaging parameters created a more generalizable model across different institutions, they wrote. The authors added that this combination will contribute to higher accuracy in neonatal skull stripping across various institutions in comparison to studies performed with a smaller and institution-specific dataset.
Read the full study here.