An automated method based on a machine-learning algorithm and MRI radiomics can differentiate between low-grade and high-grade gliomas, according to research presented at the annual Society for Imaging Informatics in Medicine (SIIM) conference in Kissimmee, FL.
After developing a workflow to support it, researchers from Yale School of Medicine created an automated approach that segments gliomas on brain MR exams, performs radiomics analysis, and then predicts if the tumor is high or low grade. In testing, their approach yielded an area under the curve (AUC) of 0.86.
“We were able to develop a PACS-based auto-segmentation tool, which was linked to a high- versus low-grade glioma prediction tool,” said Sara Merkaj, a postgraduate research fellow. “This algorithm could potentially be incorporated into clinical practice.”
Gliomas are the most common primary brain tumors and are classified -- based on pathology and molecular markers -- as grades 1-4 under the World Health Organization (WHO) criteria. Grades 1-2 are considered low-grade gliomas, while grades 3-4 are deemed to be high grades.
“There’s a difference in prognosis and treatment between high- and low-grade gliomas, and the current gold standard for grading is biopsy, which is lengthy, invasive, and associated with certain surgical needs,” she said. “For that reason, there is a critical need for noninvasive, preoperative glioma prediction tools.”
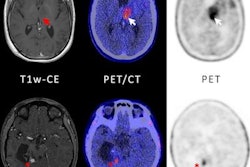
To develop their model, the researchers used the dataset from the Brain Tumor Segmentation (BraTS) 2021 challenge to pretrain a U-Net algorithm to perform auto-segmentation of whole, core, and necrotic segments of primary brain tumors on fluid-attenuated inversion recovery (FLAIR) MR images. They then validated the model using pre-therapy MRI scans from patients at Yale New Haven Health.
After nearly 2,000 radiomics features were extracted using the PyRadiomics software from three regions of interest and six sequences, an XGBoost machine-learning algorithm was then trained to classify high- versus low-grade gliomas. The researchers internally validated the model using 30 repetitions of five-fold cross-validation.
Furthermore, the researchers employed a UNet Transformer deep-learning architecture to embed their algorithm into PACS software (Visage 7, Visage Imaging). The PyRadiomics software was also embedded within Visage 7 to extract radiomics features from the image segmentations.
In testing that included 324 high- and low-grade gliomas, the algorithm yielded a mean AUC of 0.86.
In future directions, the researchers are currently exploring new methods for dealing with missing data and imbalanced datasets, according to Merkaj. In addition, they’re planning to test this algorithm in a prospective clinical trial, with the goal of later incorporating it into clinical practice, she said.
What’s more, they’re currently exploring the use of deep-learning algorithms for glioma prediction. In addition, they would like to combine predictions of tumor grade with molecular subtypes, especially isocitrate dehydrogenase (IDH) status, Merkaj said.