An AI algorithm can generate synthetic 3-tesla images from MRI data acquired on a portable 64-mT MRI scanner from patients with multiple sclerosis (MS), according to research published April 22 in Radiology.
Called LowGAN, the generative adversarial network (GAN) yielded both qualitative and quantitative improvement for the low-field-strength MRI scans, a team led by first author Alfredo Lucas of the University of Pennsylvania reported. The researchers said that their results serve as preliminary evidence for the feasibility of combining portable MRI with a deep-learning algorithm is feasible for screening, monitoring, and characterizing MS.
“For portable MRI in multiple sclerosis (MS), LowGAN produced 3T-like images, recovered regional brain volumes, and increased white matter lesion conspicuity,” the authors wrote.
Although portable low-field-strength MRI scanners show promise for increasing access to neuroimaging for both clinical and research purposes, these systems produce lower-quality images than conventional high-field-strength scanners, according to the researchers. As a result, the group sought to develop and test a deep learning-based architecture to generate synthetic high field strength from the data acquired on these portable systems.
They developed LowGAN on a set of 50 patients who had received same-day brain MRI scans with T1-weighted, T2-weighted, and fluid-attenuated inversion recovery (FLAIR) imaging sequences on both a Swoop MRI scanner (Hyperfine) and a 3-tesla MRI scanner (Siemens Healthineers). LowGAN was then validated on a separate set of 13 patients, which included four without MS.
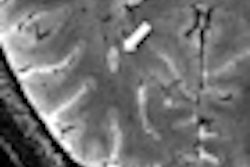
 Figure: Improved image quality with a generative adversarial network architecture for low- to high-field-strength image translation, called LowGAN. (A–C) Top: High-field-strength (3-T) and low-field-strength (64-mT) images and LowGAN outputs for a single participant across (A) fluid-attenuated inversion recovery (FLAIR), (B) T1-weighted (T1w), and (C) T2-weighted (T2w) contrasts. Bottom: Graphs demonstrate the structural similarity index (SSIM), peak signal-to-noise ratio (PSNR), normalized cross-correlation (NCC), and feature similarity index (FSIM) between high field strength and low field strength (64-mT) and high field strength and LowGAN outputs. a.u. = arbitrary units.****p ≤ 0.0001. Images and caption courtesy of Radiology and the RSNA.
Figure: Improved image quality with a generative adversarial network architecture for low- to high-field-strength image translation, called LowGAN. (A–C) Top: High-field-strength (3-T) and low-field-strength (64-mT) images and LowGAN outputs for a single participant across (A) fluid-attenuated inversion recovery (FLAIR), (B) T1-weighted (T1w), and (C) T2-weighted (T2w) contrasts. Bottom: Graphs demonstrate the structural similarity index (SSIM), peak signal-to-noise ratio (PSNR), normalized cross-correlation (NCC), and feature similarity index (FSIM) between high field strength and low field strength (64-mT) and high field strength and LowGAN outputs. a.u. = arbitrary units.****p ≤ 0.0001. Images and caption courtesy of Radiology and the RSNA.
On the validation set, LowGAN yielded an improvement in image quality for the ultralow-field-strength images, as calculated by a statistically significant improvement in structural similarity index for FLAIR images (0.85 to 0.88, p < 0.001).
What’s more, LowGAN led to higher segmentation agreement for white-matter lesions with the actual 3-tesla scans. These results (Dice score from 0.28 to 0.32, p < 0.001) point toward increased lesion conspicuity, according to the researchers.
In addition, the model enhanced morphometric measurements; volumetric differences between synthesized and actual high-field-strength images for major anatomic structures were reduced to a statistically insignificant level 483.6 cm3 vs. 482.1 cm3 (p = 0.99).
The authors also noted that their approach serves as a model for detecting brain lesions and atrophy at low field strength and could be applied to other disease processes.
“Future research should focus on validating LowGAN across larger MS cohorts, different neurologic conditions, integrating it into clinical workflows, and exploring transfer learning to enhance its generalizability,” the authors concluded.
In an accompanying editorial, Shuncong Wang, MD, PhD, of the University of Cambridge in the U.K., and Greg Zaharchuk, MD, PhD, of Stanford University noted that challenges remain for the translational use of LowGAN in clinical care.
“Although the results are promising, work is clearly needed to ensure a robust and clinically translatable performance, which could ultimately enhance diagnostic accuracy and improve care for patients with MS,” they wrote.
The full study and accompanying editorial can be found here and here.