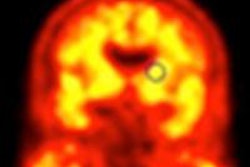
A computer-aided detection (CAD) algorithm that makes use of anatomic data from MRI scans can help radiologists differentiate between benign and malignant brain tumors on FDG-PET images, according to researchers from Yamaguchi University in Japan.
Using an internally developed algorithm that classifies tumors via image texture features on FDG-PET, a team led by Dr. Shoji Kido, PhD, found that the CAD tool yielded 100% sensitivity and 80% specificity in a 24-patient study. It also outperformed classification based on the commonly used FDG-PET quantification technique of maximum lesion standardized uptake value (SUVmax).
"The performance of our texture-based method was significantly superior to that of the SUVmax-based method," he said.
Kido presented the research during a scientific session at RSNA 2013 in Chicago.
Differentiating between malignant and benign brain tumors is important for clinical management and strategy. To help with differential diagnosis on F-18 FDG PET brain images, quantification of SUVmax with the use of optimal cutoff values has been adopted.
"However, the optimal cutoff value for differential diagnosis has significant overlap of SUV between benign and malignant lesions," he said.
As a result, the Yamaguchi team sought to develop a CAD method that utilizes texture analysis and a pattern classification technique to analyze the FDG uptake distribution of brain tumors to assist radiologists in differentiating benign from malignant lesions.
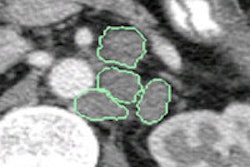
Because it can be difficult to determine the contours of tumors on PET, the technique uses MRI as the basis for segmentation of tumor regions on PET images. After patients receive MR followed by PET imaging, manual segmentation of the tumor region on MR images is performed.
Next, registration of MR and PET images is achieved by superimposing the images using a 3D nonsolid registration algorithm. The tumor region on the PET images is segmented based on the MRI tumor segmentation results.
Texture features representing FDG uptake distributions are then obtained from these tumor regions, and four optimal parameters to distinguish between benign and malignant lesions are chosen, according to the researchers. Finally, pattern classification is performed via the LibSVM open-source support vector machine (SVM) machine-learning libraries, Kido said.
The researchers retrospectively tested their technique on 24 patients who received both PET and MRI studies. Kido and colleagues compared the results with those obtained via a classification based on SUVmax with a threshold cutoff value of 8.8.
Of the 24 patients, 10 (3 men, 7 women; average age, 54.4 ± 11.4 years) had benign lesions, all of which were meningioma. The remaining 14 patients (7 men, 7 women; average age, 66.6 ± 10 years) had six glioblastomas, four malignant lymphomas, three metastases, and one oligodendroglioma.
| CAD vs. SUVmax for lesion classification | ||
| Measure | SUVmax-based lesion classification | CAD software |
| Sensitivity | 92.9% | 100% |
| Specificity | 20% | 80% |
| Accuracy | 62.5% | 91.7% |
The difference in performance between the two methods was statistically significant (p < 0.05).
"The texture-based method indicated high performance compared with the SUVmax-based method," Kido said.
A prospective study is now required for further improvement of the CAD tool, Kido noted.