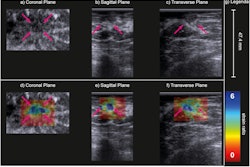
A deep-learning method could improve clinical strategies for addressing ultrasound BI-RADS 4A lesions, a study published February 20 in Clinical Breast Cancer found.
Researchers led by Mei Yi from Southern Medical University in Guangzhou, China, found that their deep learning model achieved higher sensitivities than those of clinical experience alone and that the performance of radiologists improved with deep-learning assistance.
“Using deep learning may guide clinicians to make precise clinical decisions and avoid overtreatment of benign lesions,” Yi and co-authors wrote.
Biopsy is typically recommended for ultrasound BI-RADS 4A lesions. However, the researchers noted that the malignancy rate of these lesions ranges from just 2% to 10%. They suggested that based on this, better assessment to distinguish benign from malignant lesions can make way for improved clinical decision-making.
Radiologists have explored the potential of deep learning to extract ultrasonic features that may be missed by imagers. While deep learning has shown its potential to improve accuracy and efficiency in breast cancer screening, the team pointed out a lack of relevant data on applying deep learning to ultrasound BI-RADS 4A lesions.
Using a convolutional neural network (CNN), Yi and colleagues developed a predictive nomogram based on extracted clinical and ultrasound features. Their goal was to see if the nomogram could improve the workup of ultrasound BI-RADS 4A lesions and avoid unnecessary biopsies. They highlighted the use of a ResNet18 neural network that used weights trained on another large 2D dataset of benign and malignant breast ultrasound.
The team included data from 1,616 women from two hospitals. The women were randomly placed into training and internal validation cohorts at a ratio of 7:3. Meanwhile, another 100 women were placed into an external validation group.
The nomogram achieved higher area under the curve (AUC) and sensitivity values than clinical experience alone with sacrificing specificity.
| Performance of deep-learning nomogram in distinguishing ultrasound BI-RADS 4A lesions | |||
|---|---|---|---|
| Clinical experience | Nomogram | p-value | |
| AUC (internal validation) | 0.863 | 0.916 | < 0.001 |
| AUC (external validation) | 0.776 | 0.884 | 0.05 |
| Sensitivity (internal validation) | 72.41% | 81.03% | 0.044 |
| Sensitivity (external validation) | 81.25% | 93.75% | 0.4795 |
| Specificity (internal validation) | 86.47% | 84.91% | 0.353 |
Using decision curves, the team also reported that the deep-learning nomogram adds more clinical net benefit.
The performance of two radiologists significantly improved with assistance from the nomogram. This included AUCs of 0.801 and 0.8 with assistance compared with 0.712 and 0.547 with no assistance, respectively. The same trend went for specificity, with the nomogram improving performance from 70.93% to 74.42% for one radiologist (p < 0.001) and from 59.3% to 81.4% (p = 0.004) for the other radiologist.
Finally, the deep-learning method decreased unnecessary biopsy rates for both radiologists by 6.7% and 24%, respectively.
The study authors highlighted that these results are suggestive of deep learning’s usefulness in clinical practice.
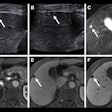
“Notably, we found that a hyperechoic halo was significantly related to breast cancer, which may be caused by the infiltration of cancerous tissue into the peripheral tissue,” they added. “Hence, we suggest that BI-RADS 4A breast lesions with hyperechoic halos should be paid high attention to, especially in older women.”
The full study can be found here.