Although automated volumetry can be performed on an ultrafast 3D brain MRI sequence, conventional image acquisition still yielded the best results in a study published online January 5 in the American Journal of Roentgenology.
A team of researchers led by corresponding author Dr. Hye Jin Baek, PhD, of Gyeongsang National University School of Medicine in Changwon, South Korea, used commercial and open-source volumetry software applications to calculate brain volume in a retrospective study involving 36 patients with memory impairment who had received 3-tesla brain MRI.
These automated volumetry methods showed a high level of agreement on both conventional T1-weighted (T1W) imaging and an isotropic ultrafast echo-planar imaging (EPI) T1-weighted sequence that was performed due to patient motion. But there were also some shortcomings from the ultrafast technique, including significant mean differences and consistent systematic biases between the two sequences.
"The ultrafast T1W sequence facilitates brain volumetry in patients with motion, though the current conventional T1W sequence remains preferred when of sufficient quality," the authors wrote.
Although volumetric segmentation of 3D T1-weighted imaging can help to detect subtle changes in individual brain regions, conventional 3D T1-weighted imaging is typically performed after contrast administration and requires 3-5 minutes of scanning time. It's also prone to motion artifacts that result in nondiagnostic exams, according to the researchers.
As a result, they sought to compare the performance of automated volumetry methods on both conventional MRI and an ultrafast sequence in 36 patients. Of these patients, 12 had subjective cognitive impairment, but they were found after the MRI scans to have normal objective cognitive function based on neuropsychiatric evaluation. Eight were diagnosed with mild cognitive impairment after MRI, while 16 were diagnosed with Alzheimer's disease after the imaging exam.
The patients received isotropic 3D T1-weighted imaging using an inversion recovery gradient-recalled echo (GRE) sequence at a slice thickness of 1 mm and acquisition time of 3 minutes and 4 seconds. Patients exhibiting motion also received an isotropic ultrafast 3D-EPI sequence with a slice thickness of 1.2 mm and an acquisition time of 30 seconds in the same scanning session.
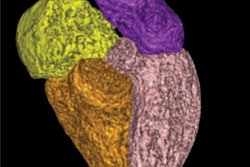
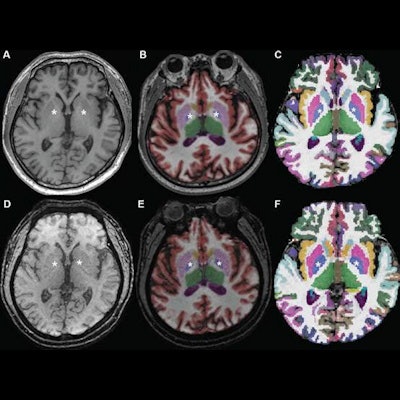
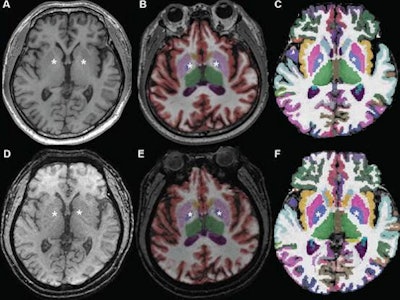 Axial MR images at basal ganglia in a 63-year-old woman with subjective cognitive impairment. Conventional 3D T1-weighted image (A) without segmentation, (B) with segmentation by Neuroquant software, and (C) with segmentation by Freesurfer software; ultrafast 3D-EPI T1-weighted image (D) without segmentation, (E) with segmentation by Neuroquant, and (F) with segmentation by Freesurfer. Pallidum (asterisks) appears larger for Freesurfer (C, F) than for NeuroQuant (B, E), but appears similar in size between two sequences for each software package. For both software packages, bilateral frontal and occipital cortices (B, C) at bone-tissue interface appear more color-coded for conventional than for ultrafast sequence, contributing to larger cortical gray matter for conventional sequence. All images and caption courtesy of the American Journal of Roentgenology.
Axial MR images at basal ganglia in a 63-year-old woman with subjective cognitive impairment. Conventional 3D T1-weighted image (A) without segmentation, (B) with segmentation by Neuroquant software, and (C) with segmentation by Freesurfer software; ultrafast 3D-EPI T1-weighted image (D) without segmentation, (E) with segmentation by Neuroquant, and (F) with segmentation by Freesurfer. Pallidum (asterisks) appears larger for Freesurfer (C, F) than for NeuroQuant (B, E), but appears similar in size between two sequences for each software package. For both software packages, bilateral frontal and occipital cortices (B, C) at bone-tissue interface appear more color-coded for conventional than for ultrafast sequence, contributing to larger cortical gray matter for conventional sequence. All images and caption courtesy of the American Journal of Roentgenology.The researchers then used both the commercial NeuroQuant software application (CorTech Labs) and FreeSurfer (Harvard University) to perform automated brain segmentation. Next, they compared the measurements between sequences for nine regions in each brain hemisphere.
"For automated brain volume measurements from ultrafast 3D-EPI T1W imaging, compared with conventional 3D T1W imaging, most regions demonstrated at least substantial agreement between the two sequences, yet also significantly different mean values, moderate or large effect sizes, consistent systematic biases with wide limits of agreement between the sequences," the authors wrote.
Intraclass correlation coefficient analysis demonstrated substantial to almost perfect agreement between the two sequences for most regions bilaterally, according to the researchers. However, they also found significant mean differences between sequences.
The authors reported, for example, that volume was significantly greater for the ultrafast sequence in the left hemisphere on the NeuroQuant software, and significantly greater for conventional imaging in three regions. In addition, the standardized effect size between sequences was moderate for four regions and large for one region. Furthermore, the mean bias -- ultrafast minus the conventional imaging -- was greatest in the cortical gray matter bilaterally: -50.6 cm3 for left and -50.02 cm3 for the right).
What's more, variation between the two sequences was found in both the subset of 16 patients with Alzheimer's disease and the 20 patients without the disease.
"Considering these factors, brain volumetry measurements obtained using different MRI sequences and different software packages should be interpreted carefully," the authors wrote.