In patients with multiple myeloma (MM), an MRI radiomics machine-learning algorithm can identify minimal residual disease (MRD) status, a biomarker linked with longer survival, according to a new study published in Clinical Radiology.
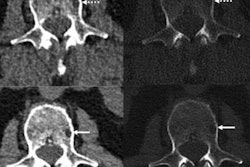
A group of researchers led by X. Xiong of The First Affiliated Hospital of Soochow University in Jiangsu, China, trained machine-learning algorithm to extract and analyze radiomics features on T1-weighted T1-weighed imaging and fat-saturated T2-weighted imaging sequences on lumbar spine MRI. In testing, the best-performing model -- a linear support vector machine (SVM) classifier -- yielded an area under the curve (AUC) for MRD status of up to 0.8.
"The linear SVM-based machine-learning method can offer a noninvasive tool for discriminating MRD status in MM," the authors wrote.
An emerging biomarker for MM, MRS is considered to represent the deepest level of treatment response for MM. Recent radiomics studies have shown potential for predicting the high-risk cytogenetic status and evaluating MM treatment response, according to the researchers. However, both of these studies had used small data sets.
In their study, the researchers sought to explore the possibility of discriminating MRD status on MRI and to identify the best machine-learning methods to optimize the clinical treatment regimen.
The researchers retrospectively gathered data from 83 newly diagnosed patients who had received whole-body MRI prior to therapy. Of these, 59 were used for model training and 24 were set aside for validation of the algorithms.
Radiomics models were developed based on T1-weighed imaging and fat-saturated T2-weighted imaging sequences using five different classifiers: random forest, K-nearest neighbor, naïve Bayes, linear SVM, and radial SVM. After logistic regression analysis was performed, bone-marrow infiltrate ratio was the only feature retained for radiomics analysis.
The researchers also trained a traditional model based on clinical data, including age, gender, International Staging System stage, fluorescence in situ hybridization (FISH) status, serum protein level, bone marrow infiltrate ratio, calcium, creatinine, and albumin. Finally, they developed a combined model incorporating analysis of both radiomics and clinical features.
In the validation set, the linear SVM-based radiomics model yielded an AUC of 0.708 on the T1-weighted images and an AUC of 0.8 on the fat-saturated T2-weighted images, outperforming the traditional model. In addition, the combined model did not yield a statistically significant difference in performance over the radiomics model.
"In summary, the present results demonstrate the performance of radiomics analysis and machine learning of MRI, which can discriminate MRD status accurately after induction treatment," the authors concluded. "As the research was retrospective, prospective studies and multiple time-point analyses are needed to further examine the ability of them method to discriminate MRD status."
The full study can be found here.