Tuberculosis (TB) can be consistently detected on lateral chest x-rays by an ensemble of two different types of deep-learning networks, according to research published online February 24 in Frontiers in Genetics.
Although a number of AI studies have shown the potential for using AI to detect TB, these algorithms typically utilize only the frontal chest x-ray projections: the posteroanterior (PA) and the anteroposterior (AP) views. However, a research team led by Sivaramakrishnan Rajaraman, PhD, of the U.S. National Institutes of Health (NIH) trained ensembles of artificial intelligence (AI) algorithms -- convolutional neural networks (CNNs) and vision transformers (ViTs) -- that were able to take advantage of the diagnostic information contained in the lateral views.
"The main contribution of this work is a systematic approach that benefits from constructing ensembles of the best models from both worlds (i.e., CNNs and ViTs) to detect TB-consistent findings using lateral [chest x-rays] through reduced prediction variance and improved performance," the authors wrote.
Lateral chest radiographs help to detect clinically suspected pulmonary TB, particularly in children. A number of studies in the literature have showed the benefit of lateral chest x-rays in, for example, identifying mediastinal or hilar lymphadenopathy and for providing information on the thoracic cage, pleura, lungs, pericardium, heart, mediastinum, and upper abdomen, according to the researchers.
"These studies underscore the importance of using lateral [chest x-ray] projections as they carry useful information on disease manifestation and progression; hence, this study aims to explore these least studied types of [chest x-ray] projection (the lateral) and propose a novel approach for detecting TB-consistent findings," the authors wrote.
As ViTs have also shown promising results in image-recognition tasks, the researchers sought to explore the potential benefit of combining both CNNs and ViTs into ensemble models. They trained and tested the individual and ensemble models using the CheXpert dataset and PadChest datasets. Of these, 90% were used for training and 10% for testing.
In the training process, researchers meant to transfer modality-specific knowledge to the algorithms and then fine-tune the models for detecting findings consistent with TB.
In testing, the ensembles yielded performances such as a Matthew's correlation coefficient (MCC) of 0.8136 and an area under the curve (AUC) as high as 0.949, significantly better than the best individual models.
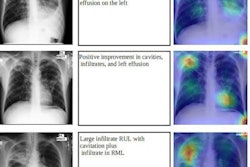
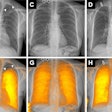
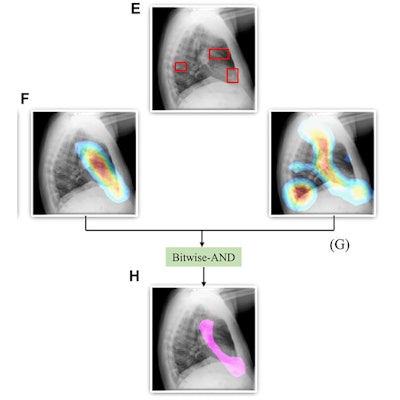
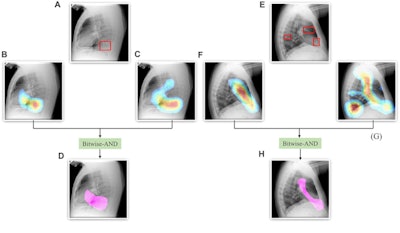 A Bitwise-AND ensemble generated using the heatmaps produced by the top two performing models, VGG-16 and DenseNet-121 models. (A) and (E) Sample test lateral chest x-rays with expert ground truth annotations (shown in red bounding box); (B) and (F) heatmaps produced by the VGG-16 model; (C) and (G) heatmaps produced by the DenseNet-121 model, and (D) and (H) mask resulting from the Bitwise-AND operation of the heatmaps produced by the VGG-16 and DenseNet-121 models. Image and caption courtesy of Sivaramakrishnan Rajaraman, PhD, et al and Frontiers in Genomics through Creative Commons Attribution 4.0 International License.
A Bitwise-AND ensemble generated using the heatmaps produced by the top two performing models, VGG-16 and DenseNet-121 models. (A) and (E) Sample test lateral chest x-rays with expert ground truth annotations (shown in red bounding box); (B) and (F) heatmaps produced by the VGG-16 model; (C) and (G) heatmaps produced by the DenseNet-121 model, and (D) and (H) mask resulting from the Bitwise-AND operation of the heatmaps produced by the VGG-16 and DenseNet-121 models. Image and caption courtesy of Sivaramakrishnan Rajaraman, PhD, et al and Frontiers in Genomics through Creative Commons Attribution 4.0 International License.Delving further into the results from the algorithms, the researchers used class-selective maps to review the decisions of the CNNs and attention maps to review the results of the ViTs. These results were then combined to highlight the regions of the image that contributed to the algorithms' final output.
The study authors found that the accuracy of the models was not related to its localization of the disease region of interest.
"This underscores the need for visualization of localized disease prediction regions to verify model credibility," they wrote.
The researchers also found that the two best-performing ensembles produced a mean average precision of 0.182, significantly better than the individual models and other ensembles.